6QYN
 
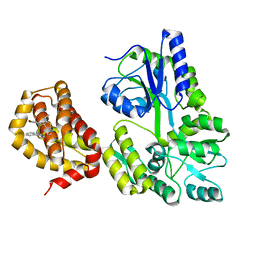 | | Structure of MBP-Mcl-1 in complex with compound 10d | | Descriptor: | (2~{R})-2-[5-(3-chloranyl-2-methyl-4-oxidanyl-phenyl)-6-ethyl-thieno[2,3-d]pyrimidin-4-yl]oxy-3-phenyl-propanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-09 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6QYP
 
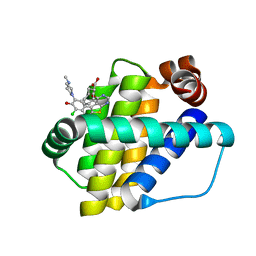 | | Structure of Mcl-1 in complex with compound 13 | | Descriptor: | (2~{R})-2-[5-[3-chloranyl-2-methyl-5-(4-methylpiperazin-1-yl)-4-oxidanyl-phenyl]-6-ethyl-thieno[2,3-d]pyrimidin-4-yl]oxy-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-09 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6QZ5
 
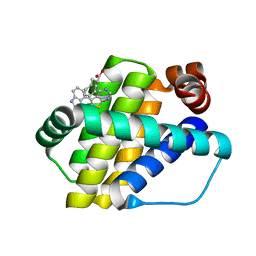 | | Structure of Mcl-1 in complex with compound 8a | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-4-yl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6QYO
 
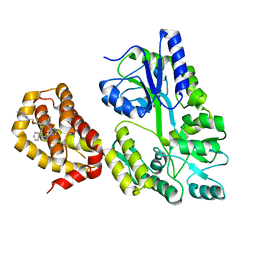 | | Structure of MBP-Mcl-1 in complex with compound 18a | | Descriptor: | (2~{R})-2-[5-[3-chloranyl-2-methyl-4-[2-(4-methylpiperazin-1-yl)ethoxy]phenyl]-6-ethyl-thieno[2,3-d]pyrimidin-4-yl]oxy-3-phenyl-propanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-09 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
1PMB
 
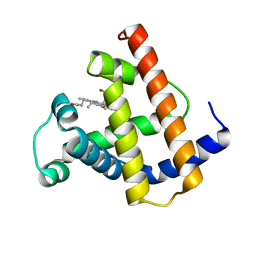 | | THE DETERMINATION OF THE CRYSTAL STRUCTURE OF RECOMBINANT PIG MYOGLOBIN BY MOLECULAR REPLACEMENT AND ITS REFINEMENT | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Smerdon, S.J, Oldfield, T.J, Dodson, E.J, Dodson, G.G, Hubbard, R.E, Wilkinson, A.J. | | Deposit date: | 1989-11-27 | | Release date: | 1990-01-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Determination of the crystal structure of recombinant pig myoglobin by molecular replacement and its refinement.
Acta Crystallogr.,Sect.B, 46, 1990
|
|
1PMI
 
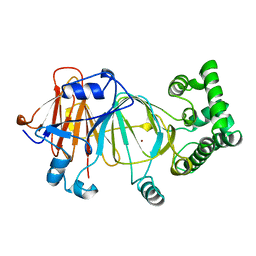 | | Candida Albicans Phosphomannose Isomerase | | Descriptor: | PHOSPHOMANNOSE ISOMERASE, ZINC ION | | Authors: | Cleasby, A, Skarzynski, T, Wonacott, A, Davies, G.J, Hubbard, R.E, Proudfoot, A.E.I, Wells, T.N.C, Payton, M.A, Bernard, A.R. | | Deposit date: | 1996-04-03 | | Release date: | 1997-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The x-ray crystal structure of phosphomannose isomerase from Candida albicans at 1.7 angstrom resolution.
Nat.Struct.Biol., 3, 1996
|
|
2YEG
 
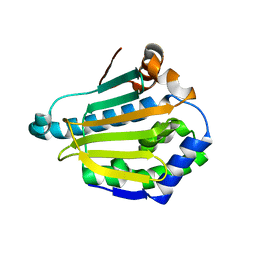 | |
2YEE
 
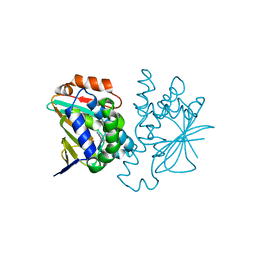 | |
2YEH
 
 | |
2YE4
 
 | |
2YEC
 
 | |
2YEB
 
 | |
2YEF
 
 | |
2XXK
 
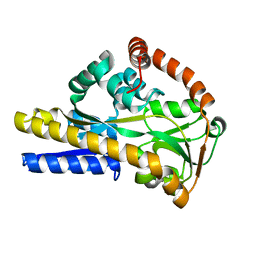 | | SiaP complex | | Descriptor: | N-acetyl-beta-neuraminic acid, SIALIC ACID-BINDING PERIPLASMIC PROTEIN SIAP | | Authors: | Fischer, M, Hubbard, R.E. | | Deposit date: | 2010-11-10 | | Release date: | 2011-11-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Siap Complex
To be Published
|
|
2XWO
 
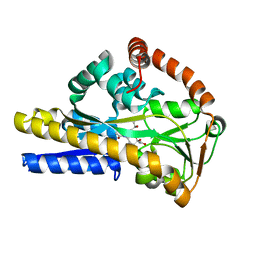 | | SiaP R147E mutant in complex with sialylamide | | Descriptor: | (2S,4S,5R,6R)-5-acetamido-2,4-dihydroxy-6-[(1R,2R)-1,2,3-trihydroxypropyl]oxane-2-carboxamide, SIALIC ACID-BINDING PERIPLASMIC PROTEIN SIAP | | Authors: | Fischer, M, Hubbard, R.E. | | Deposit date: | 2010-11-04 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Tripartite ATP-Independent Periplasmic (Trap) Transporters Use an Arginine-Mediated Selectivity Filter for High Affinity Substrate Binding.
J.Biol.Chem., 290, 2015
|
|
2XWV
 
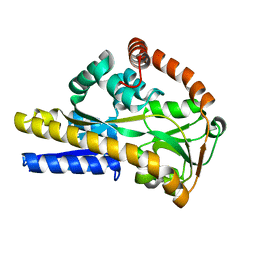 | | SiaP complex | | Descriptor: | N-acetyl-beta-neuraminic acid, SIALIC ACID-BINDING PERIPLASMIC PROTEIN SIAP | | Authors: | Fischer, M, Hubbard, R.E. | | Deposit date: | 2010-11-05 | | Release date: | 2011-11-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Tripartite ATP-Independent Periplasmic (Trap) Transporters Use an Arginine-Mediated Selectivity Filter for High Affinity Substrate Binding.
J.Biol.Chem., 290, 2015
|
|
2YEA
 
 | |
2YED
 
 | |
2YE8
 
 | |
2YE2
 
 | | HSP90 inhibitors and drugs from fragment and virtual screening | | Descriptor: | HEAT SHOCK PROTEIN HSP 90-ALPHA, METHYL 2-(3,5-DIHYDROXYPHENYL)ETHANOATE, SULFATE ION | | Authors: | Roughley, S.D, Hubbard, R.E, Baker, L.M. | | Deposit date: | 2011-03-25 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | How Well Can Fragments Explore Accessed Chemical Space? a Case Study from Heat Shock Protein 90.
J.Med.Chem., 54, 2011
|
|
2YEI
 
 | | HSP90 inhibitors and drugs from fragment and virtual screening | | Descriptor: | HEAT SHOCK PROTEIN HSP 90-ALPHA, METHYL 2-(3,5-DIHYDROXYPHENYL)ETHANOATE, METHYL 5-ISOXAZOL-5-YL-3-METHYL-1H-PYRAZOLE-4-CARBOXYLATE | | Authors: | Roughley, S.D, Hubbard, R.E, Baker, L.M. | | Deposit date: | 2011-03-25 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | How Well Can Fragments Explore Accessed Chemical Space? a Case Study from Heat Shock Protein 90.
J.Med.Chem., 54, 2011
|
|
2XWK
 
 | | SiaP R147A mutant in complex with Neu5Ac | | Descriptor: | N-acetyl-beta-neuraminic acid, SIALIC ACID-BINDING PERIPLASMIC PROTEIN SIAP | | Authors: | Fischer, M, Hubbard, R.E. | | Deposit date: | 2010-11-04 | | Release date: | 2011-11-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Tripartite ATP-Independent Periplasmic (Trap) Transporters Use an Arginine-Mediated Selectivity Filter for High Affinity Substrate Binding.
J.Biol.Chem., 290, 2015
|
|
2XWI
 
 | | SiaP R147K mutant in complex with Neu5Ac | | Descriptor: | N-acetyl-beta-neuraminic acid, SIALIC ACID-BINDING PERIPLASMIC PROTEIN SIAP | | Authors: | Fischer, M, Hubbard, R.E. | | Deposit date: | 2010-11-04 | | Release date: | 2011-11-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.195 Å) | | Cite: | Tripartite ATP-Independent Periplasmic (Trap) Transporters Use an Arginine-Mediated Selectivity Filter for High Affinity Substrate Binding.
J.Biol.Chem., 290, 2015
|
|
2YE5
 
 | |
2YE7
 
 | | HSP90 inhibitors and drugs from fragment and virtual screening | | Descriptor: | 2-[(3,5-DIMETHYL-1H-PYRAZOL-4-YL)SULFANYL]-5-METHYLSULFANYL-1,3,4-THIADIAZOLE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Roughley, S.D, Hubbard, R.E, Baker, L.M. | | Deposit date: | 2011-03-25 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | How Well Can Fragments Explore Accessed Chemical Space? a Case Study from Heat Shock Protein 90.
J.Med.Chem., 54, 2011
|
|
