7LUG
 
 | | Crystal structure of the pnRFP B30Y mutant | | Descriptor: | PHOSPHATE ION, Red Fluorescent pnRFP B30Y mutant | | Authors: | Huang, M, Ng, H.L, Pang, Y, Zhang, S, Fan, Y, Yeh, H, Xiong, Y, Li, X, Ai, H. | | Deposit date: | 2021-02-22 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Development, Characterization, and Structural Analysis of a Genetically Encoded Red Fluorescent Peroxynitrite Biosensor
To Be Published
|
|
7LQO
 
 | | Crystal structure of a genetically encoded red fluorescent peroxynitrite biosensor, pnRFP | | Descriptor: | PHOSPHATE ION, red fluorescent peroxynitrite biosensor pnRFP | | Authors: | Huang, M, Ng, H.L, Pang, Y, Zhang, S, Fan, Y, Yeh, H, Xiong, Y, Li, X, Ai, H. | | Deposit date: | 2021-02-14 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Development, Characterization, and Structural Analysis of a Genetically Encoded Red Fluorescent Peroxynitrite Biosensor
To Be Published
|
|
7CRW
 
 | | Cryo-EM structure of rNLRP1-rDPP9 complex | | Descriptor: | Dipeptidyl peptidase 9, NLR family protein 1 | | Authors: | Huang, M.H, Zhang, X.X, Wang, J, Chai, J.J. | | Deposit date: | 2020-08-14 | | Release date: | 2021-03-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural and biochemical mechanisms of NLRP1 inhibition by DPP9.
Nature, 592, 2021
|
|
7CRV
 
 | |
1AHW
 
 | | A COMPLEX OF EXTRACELLULAR DOMAIN OF TISSUE FACTOR WITH AN INHIBITORY FAB (5G9) | | Descriptor: | IMMUNOGLOBULIN FAB 5G9 (HEAVY CHAIN), IMMUNOGLOBULIN FAB 5G9 (LIGHT CHAIN), TISSUE FACTOR | | Authors: | Huang, M, Syed, R, Stura, E.A, Stone, M.J, Stefanko, R.S, Ruf, W, Edgington, T.S, Wilson, I.A. | | Deposit date: | 1997-04-10 | | Release date: | 1998-02-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The mechanism of an inhibitory antibody on TF-initiated blood coagulation revealed by the crystal structures of human tissue factor, Fab 5G9 and TF.5G9 complex.
J.Mol.Biol., 275, 1998
|
|
2PQS
 
 | | Crystal Structure of the Bovine Lactadherin C2 Domain | | Descriptor: | Lactadherin | | Authors: | Huang, M, Furie, B.C. | | Deposit date: | 2007-05-02 | | Release date: | 2007-08-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the bovine lactadherin C2 domain, a membrane binding motif, shows similarity to the C2 domains of factor V and factor VIII.
J.Mol.Biol., 371, 2007
|
|
6ION
 
 | | The complex of C4.4A with its antibody 11H10 Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ly6/PLAUR domain-containing protein 3, ... | | Authors: | Huang, M.D, Jiang, Y.B, Yuan, C, Lin, L. | | Deposit date: | 2018-10-30 | | Release date: | 2020-01-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structures of Human C4.4A Reveal the Unique Association of Ly6/uPAR/alpha-neurotoxin Domain
Int J Biol Sci, 16, 2020
|
|
6IOM
 
 | | Crystal structure of human C4.4A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ly6/PLAUR domain-containing protein 3 | | Authors: | Huang, M.D, Jiang, Y.B, Yuan, C, Lin, L. | | Deposit date: | 2018-10-30 | | Release date: | 2020-01-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Crystal Structures of Human C4.4A Reveal the Unique Association of Ly6/uPAR/alpha-neurotoxin Domain
Int J Biol Sci, 16, 2020
|
|
7W2T
 
 | | Crystal structure of TxGH116 E730Q mutant from Thermoanaerobacterium xylanolyticum with glucose | | Descriptor: | CALCIUM ION, GLYCEROL, Glucosylceramidase, ... | | Authors: | Huang, M, Pengthaisong, S, Charoenwattanasatien, R, Jitonnom, J, Ketudat Cairns, J.R. | | Deposit date: | 2021-11-24 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Systematic Functional and Computational Analysis of Glucose-Binding Residues in Glycoside Hydrolase Family GH116.
Catalysts, 12, 2022
|
|
7W2S
 
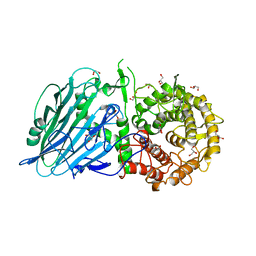 | | Crystal structure of TxGH116 E730A mutant from Thermoanaerobacterium xylanolyticum with glucose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Huang, M, Pengthaisong, S, Charoenwattanasatien, R, Jitonnom, J, Ketudat Cairns, J.R. | | Deposit date: | 2021-11-24 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Systematic Functional and Computational Analysis of Glucose-Binding Residues in Glycoside Hydrolase Family GH116.
Catalysts, 12, 2022
|
|
7W2W
 
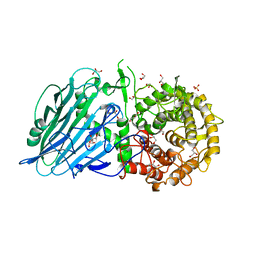 | | Crystal structure of TxGH116 R786K mutant from Thermoanaerobacterium xylanolyticum with glucose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Huang, M, Pengthaisong, S, Charoenwattanasatien, R, Jitonnom, J, Ketudat Cairns, J.R. | | Deposit date: | 2021-11-24 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Systematic Functional and Computational Analysis of Glucose-Binding Residues in Glycoside Hydrolase Family GH116.
Catalysts, 12, 2022
|
|
7W2V
 
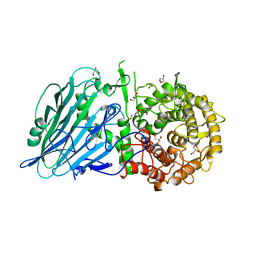 | | Crystal structure of TxGH116 R786A mutant from Thermoanaerobacterium xylanolyticum with glucose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Huang, M, Pengthaisong, S, Charoenwattanasatien, R, Jitonnom, J, Ketudat Cairns, J.R. | | Deposit date: | 2021-11-24 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Systematic Functional and Computational Analysis of Glucose-Binding Residues in Glycoside Hydrolase Family GH116.
Catalysts, 12, 2022
|
|
7W2X
 
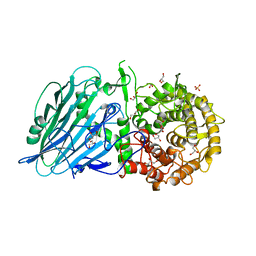 | | Crystal structure of TxGH116 R786K mutant from Thermoanaerobacterium xylanolyticum | | Descriptor: | CALCIUM ION, GLYCEROL, Glucosylceramidase, ... | | Authors: | Huang, M, Pengthaisong, S, Charoenwattanasatien, R, Jitonnom, J, Ketudat Cairns, J.R. | | Deposit date: | 2021-11-24 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Systematic Functional and Computational Analysis of Glucose-Binding Residues in Glycoside Hydrolase Family GH116.
Catalysts, 12, 2022
|
|
7X1M
 
 | | The complex structure of Omicron BA.1 RBD with BD604, S309,and S304 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, BD-604 Fab heavy chain, BD-604 Fab light chain, ... | | Authors: | Huang, M, Xie, Y.F, Qi, J.X. | | Deposit date: | 2022-02-24 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Atlas of currently available human neutralizing antibodies against SARS-CoV-2 and escape by Omicron sub-variants BA.1/BA.1.1/BA.2/BA.3.
Immunity, 55, 2022
|
|
3HU6
 
 | |
3HVE
 
 | |
3HTM
 
 | |
3HQI
 
 | |
3HQH
 
 | |
3HSV
 
 | | Structures of SPOP-Substrate Complexes: Insights into Molecular Architectures of BTB-Cul3 Ubiquitin Ligases: SPOPMATHx-MacroH2ASBCpep2 | | Descriptor: | Core histone macro-H2A.1, SULFATE ION, Speckle-type POZ protein, ... | | Authors: | Zhuang, M, Schulman, B.A, Miller, D. | | Deposit date: | 2009-06-10 | | Release date: | 2009-10-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structures of SPOP-substrate complexes: insights into molecular architectures of BTB-Cul3 ubiquitin ligases.
Mol.Cell, 36, 2009
|
|
3HQM
 
 | |
3HQL
 
 | |
1MWA
 
 | | 2C/H-2KBM3/DEV8 ALLOGENEIC COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2C T CELL RECEPTOR ALPHA CHAIN, ... | | Authors: | Luz, J.G, Huang, M.D, Garcia, K.C, Rudolph, M.G, Teyton, L, Wilson, I.A. | | Deposit date: | 2002-09-27 | | Release date: | 2002-11-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural comparison of allogeneic and syngeneic T cell receptor-peptide-major histocompatibility
complex complexes: a buried alloreactive mutation subtly alters peptide presentation substantially
increasing V(beta) Interactions.
J.EXP.MED., 195, 2002
|
|
7Q4M
 
 | | Type II beta-amyloid 42 Filaments from Human Brain | | Descriptor: | Amyloid-beta precursor protein, UNKNOWN ATOM OR ION | | Authors: | Yang, Y, Arseni, D, Zhang, W, Huang, M, Lovestam, S.K.A, Schweighauser, M, Kotecha, A, Murzin, A.G, Peak-Chew, S.Y, Macdonald, J, Lavenir, I, Garringer, H.J, Gelpi, E, Newell, K.L, Kovacs, G.G, Vidal, R, Ghetti, B, Falcon, B, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2021-11-01 | | Release date: | 2021-11-24 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of amyloid-beta 42 filaments from human brains.
Science, 375, 2022
|
|
7Q4B
 
 | | Type I beta-amyloid 42 Filaments from Human Brain | | Descriptor: | Amyloid-beta precursor protein, UNKNOWN ATOM OR ION | | Authors: | Yang, Y, Arseni, D, Zhang, W, Huang, M, Lovestam, S.K.A, Schweighauser, M, Kotecha, A, Murzin, A.G, Peak-Chew, S.Y, Macdonald, J, Lavenir, I, Garringer, H.J, Gelpi, E, Newell, K.L, Kovacs, G.G, Vidal, R, Ghetti, B, Falcon, B, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2021-10-30 | | Release date: | 2021-11-24 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structures of amyloid-beta 42 filaments from human brains.
Science, 375, 2022
|
|
