7QMN
 
 | | Endothiapepsin in complex with compound TL00150 at room-temperature (temperature ramping down structure 15) | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.Y, Aumonier, S, Wang, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Probing ligand binding of endothiapepsin by `temperature-resolved' macromolecular crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QMO
 
 | | Endothiapepsin in complex with compound TL00150 at room-temperature (temperature ramping down structure 16) | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.Y, Aumonier, S, Wang, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Probing ligand binding of endothiapepsin by `temperature-resolved' macromolecular crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QMP
 
 | | Endothiapepsin in complex with compound TL00150 at room-temperature (temperature ramping down structure 17) | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.Y, Aumonier, S, Wang, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Probing ligand binding of endothiapepsin by `temperature-resolved' macromolecular crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QMQ
 
 | | Endothiapepsin in complex with compound TL00150 at room-temperature (temperature ramping down structure 18) | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.Y, Aumonier, S, Wang, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Probing ligand binding of endothiapepsin by `temperature-resolved' macromolecular crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QMR
 
 | | Endothiapepsin in complex with compound TL00150 at room-temperature (temperature ramping up structure 1) | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.Y, Aumonier, S, Wang, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Probing ligand binding of endothiapepsin by `temperature-resolved' macromolecular crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QMT
 
 | | Endothiapepsin in complex with compound TL00150 at room-temperature (temperature ramping up structure 3) | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.Y, Aumonier, S, Wang, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Probing ligand binding of endothiapepsin by `temperature-resolved' macromolecular crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QMS
 
 | | Endothiapepsin in complex with compound TL00150 at room-temperature (temperature ramping up structure 2) | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.Y, Aumonier, S, Wang, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Probing ligand binding of endothiapepsin by `temperature-resolved' macromolecular crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QMU
 
 | | Endothiapepsin in complex with compound TL00150 at room-temperature (temperature ramping up structure 4) | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.Y, Aumonier, S, Wang, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Probing ligand binding of endothiapepsin by `temperature-resolved' macromolecular crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QMV
 
 | | Endothiapepsin in complex with compound TL00150 at room-temperature (temperature ramping up structure 5) | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.Y, Aumonier, S, Wang, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Probing ligand binding of endothiapepsin by `temperature-resolved' macromolecular crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QMW
 
 | | Endothiapepsin in complex with compound TL00150 at room-temperature (temperature ramping up structure 6) | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.Y, Aumonier, S, Wang, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Probing ligand binding of endothiapepsin by `temperature-resolved' macromolecular crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QMX
 
 | | Endothiapepsin in complex with compound TL00150 at room-temperature (temperature ramping up structure 7) | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.Y, Aumonier, S, Wang, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Probing ligand binding of endothiapepsin by `temperature-resolved' macromolecular crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QN0
 
 | | Endothiapepsin in complex with compound TL00150 at room-temperature (temperature ramping up structure 10) | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.Y, Aumonier, S, Wang, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Probing ligand binding of endothiapepsin by `temperature-resolved' macromolecular crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QN2
 
 | | Endothiapepsin in complex with compound TL00150 at room-temperature (temperature ramping up structure 12) | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.Y, Aumonier, S, Wang, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Probing ligand binding of endothiapepsin by `temperature-resolved' macromolecular crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
4QNN
 
 | |
4APV
 
 | | The Klebsiella pneumoniae primosomal PriB protein: identification, crystal structure, and ssDNA binding mode | | Descriptor: | PRIMOSOMAL REPLICATION PROTEIN N | | Authors: | Lo, Y.H, Huang, Y.H, Hsiao, C.D, Huang, C.Y. | | Deposit date: | 2012-04-06 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | Crystal Structure and DNA-Binding Mode of Klebsiella Pneumoniae Primosomal Prib Protein.
Genes Cells, 17, 2012
|
|
6L0B
 
 | | Crystal structure of dihydroorotase in complex with fluorouracil from Saccharomyces cerevisiae | | Descriptor: | 5-FLUOROURACIL, Dihydroorotase, ZINC ION | | Authors: | Guan, H.H, Huang, Y.H, Huang, C.Y, Chen, C.J. | | Deposit date: | 2019-09-26 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the interaction modes of dihydroorotase with the anticancer drugs 5-fluorouracil and 5-aminouracil.
Biochem.Biophys.Res.Commun., 551, 2021
|
|
6L0G
 
 | | Crystal structure of dihydroorotase in complex with malate at pH6 from Saccharomyces cerevisiae | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Dihydroorotase, ZINC ION | | Authors: | Guan, H.H, Huang, Y.H, Huang, C.Y, Chen, C.J. | | Deposit date: | 2019-09-26 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.053 Å) | | Cite: | Structural basis for the interaction modes of dihydroorotase with the anticancer drugs 5-fluorouracil and 5-aminouracil.
Biochem.Biophys.Res.Commun., 551, 2021
|
|
6L0A
 
 | | Crystal structure of dihydroorotase in complex with malate at pH7 from Saccharomyces cerevisiae | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Dihydroorotase, ZINC ION | | Authors: | Guan, H.H, Huang, Y.H, Huang, C.Y, Chen, C.J. | | Deposit date: | 2019-09-26 | | Release date: | 2020-12-02 | | Last modified: | 2021-12-15 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Analysis of Saccharomyces cerevisiae Dihydroorotase Reveals Molecular Insights into the Tetramerization Mechanism
Molecules, 2021
|
|
6EDQ
 
 | | Crystal Structure of the Light-Gated Anion Channelrhodopsin GtACR1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Anion channelrhodopsin 1, GLYCEROL | | Authors: | Li, H, Huang, C.Y, Wang, M, Zheng, L, Spudich, J.L. | | Deposit date: | 2018-08-10 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a natural light-gated anion channelrhodopsin.
Elife, 8, 2019
|
|
6QZH
 
 | | Structure of the human CC Chemokine Receptor 7 in complex with the intracellular allosteric antagonist Cmp2105 and the insertion protein Sialidase NanA | | Descriptor: | 3-[[4-[[(1~{R})-2,2-dimethyl-1-(5-methylfuran-2-yl)propyl]amino]-1,1-bis(oxidanylidene)-1,2,5-thiadiazol-3-yl]amino]-~{N},~{N},6-trimethyl-2-oxidanyl-benzamide, C-C chemokine receptor type 7,Sialidase A,C-C chemokine receptor type 7, D(-)-TARTARIC ACID, ... | | Authors: | Jaeger, K, Bruenle, S, Weinert, T, Guba, W, Muehle, J, Miyazaki, T, Weber, M, Furrer, A, Haenggi, N, Tetaz, T, Huang, C.Y, Mattle, D, Vonach, J.M, Gast, A, Kuglstatter, A, Rudolph, M.G, Nogly, P, Benz, J, Dawson, R.J.P, Standfuss, J. | | Deposit date: | 2019-03-11 | | Release date: | 2019-09-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Allosteric Ligand Recognition in the Human CC Chemokine Receptor 7.
Cell, 178, 2019
|
|
8WQ9
 
 | |
7D8J
 
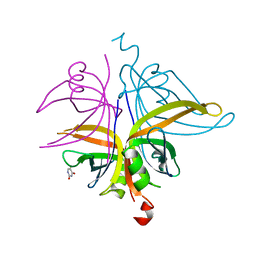 | | S. aureus SsbB with 5-FU | | Descriptor: | 5-FLUOROURACIL, Single-stranded DNA-binding protein | | Authors: | Lin, E.S, Huang, Y.H, Huang, C.Y. | | Deposit date: | 2020-10-08 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Crystal structure of the single-stranded DNA-binding protein SsbB in complex with the anticancer drug 5-fluorouracil: Extension of the 5-fluorouracil interactome to include the oligonucleotide/oligosaccharide-binding fold protein
Biochem.Biophys.Res.Commun., 534, 2020
|
|
3FWL
 
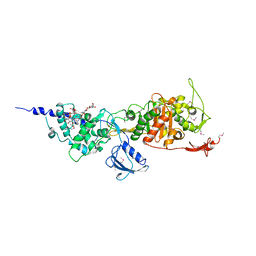 | | Crystal Structure of the Full-Length Transglycosylase PBP1b from Escherichia coli | | Descriptor: | MOENOMYCIN, Penicillin-binding protein 1B | | Authors: | Sung, M.T, Lai, Y.T, Huang, C.Y, Chou, L.Y, Wong, C.H, Ma, C. | | Deposit date: | 2009-01-19 | | Release date: | 2009-06-02 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (3.086 Å) | | Cite: | Crystal structure of the membrane-bound bifunctional transglycosylase PBP1b from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5E5C
 
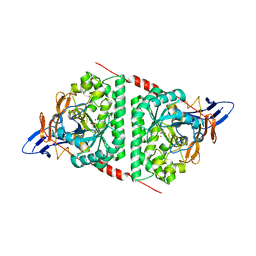 | | Crystal structure of dihydropyrimidinase from Pseudomonas aeruginosa PAO1 | | Descriptor: | D-hydantoinase/dihydropyrimidinase, ZINC ION | | Authors: | Huang, C.C, Huang, Y.H, Hsieh, Y.C, Tzeng, C.T, Chen, C.J, Huang, C.Y. | | Deposit date: | 2015-10-08 | | Release date: | 2016-09-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of dihydropyrimidinase from Pseudomonas aeruginosa PAO1: Insights into the molecular basis of formation of a dimer
Biochem.Biophys.Res.Commun., 478, 2016
|
|
6AJD
 
 | |
