8IZ8
 
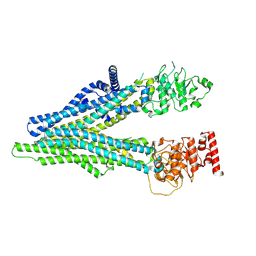 | | cryo EM structure of apo hMRP4 | | Descriptor: | ATP-binding cassette sub-family C member 4 | | Authors: | Liu, Z.M, Huang, Y. | | Deposit date: | 2023-04-06 | | Release date: | 2024-04-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural basis for substrate and inhibitor recognition of human multidrug transporter MRP4.
Commun Biol, 6, 2023
|
|
8IZ7
 
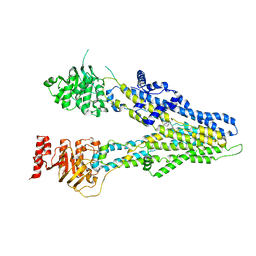 | | cryo-EM structure of sulindac-bound hMRP4 | | Descriptor: | ATP-binding cassette sub-family C member 4, [(1Z)-5-fluoro-2-methyl-1-{4-[methylsulfinyl]benzylidene}-1H-inden-3-yl]acetic acid | | Authors: | Liu, Z.M, Huang, Y. | | Deposit date: | 2023-04-06 | | Release date: | 2024-04-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for substrate and inhibitor recognition of human multidrug transporter MRP4.
Commun Biol, 6, 2023
|
|
8WI0
 
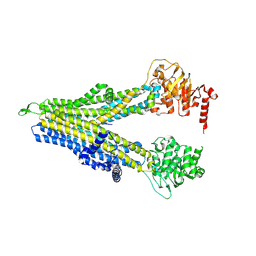 | | wt-hMRP5 inward-open | | Descriptor: | ATP-binding cassette sub-family C member 5 | | Authors: | Liu, Z.M, Huang, Y. | | Deposit date: | 2023-09-24 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Inhibition and transport mechanisms of the ABC transporter hMRP5.
Nat Commun, 15, 2024
|
|
8WI2
 
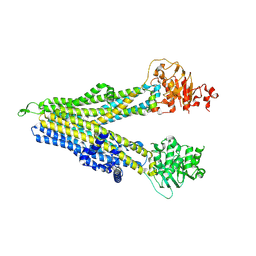 | | RD-hMRP5-inward open | | Descriptor: | ATP-binding cassette sub-family C member 5, CYS-GLN-ASP-ALA-LEU-GLU-THR-ALA-ALA-ARG-ALA-GLU-GLY-LEU-SER-LEU-ASP-ALA-SER | | Authors: | Liu, Z.M, Huang, Y. | | Deposit date: | 2023-09-24 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.06 Å) | | Cite: | Inhibition and transport mechanisms of the ABC transporter hMRP5.
Nat Commun, 15, 2024
|
|
8WI5
 
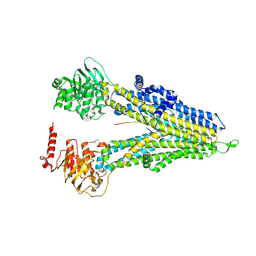 | | M5PI-bound hMRP5 | | Descriptor: | ATP-binding cassette sub-family C member 5 | | Authors: | Liu, Z.M, Huang, Y. | | Deposit date: | 2023-09-24 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Inhibition and transport mechanisms of the ABC transporter hMRP5.
Nat Commun, 15, 2024
|
|
8WI3
 
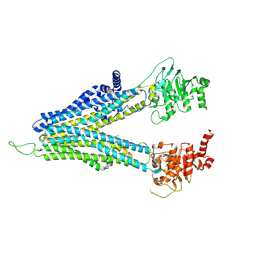 | | ND-hMRP5-inward open | | Descriptor: | ATP-binding cassette sub-family C member 5 | | Authors: | Liu, Z.M, Huang, Y. | | Deposit date: | 2023-09-24 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Inhibition and transport mechanisms of the ABC transporter hMRP5.
Nat Commun, 15, 2024
|
|
8WI4
 
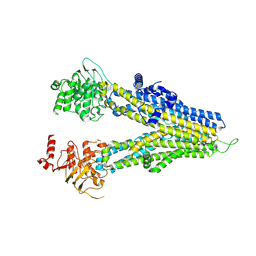 | | m6-hMRP5 inward open | | Descriptor: | ATP-binding cassette sub-family C member 5 | | Authors: | Liu, Z.M, Huang, Y. | | Deposit date: | 2023-09-24 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Inhibition and transport mechanisms of the ABC transporter hMRP5.
Nat Commun, 15, 2024
|
|
8X98
 
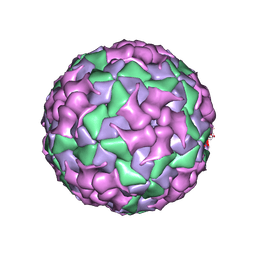 | | Cryo-EM structure of coxsackievirus A16 mature virion in complex with Fab h1A6.2 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Jiang, Y, Huang, Y, Zhu, R, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-11-29 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Cryo-EM structure of coxsackievirus A16 mature virion in complex with Fab h1A6.2
To Be Published
|
|
8X96
 
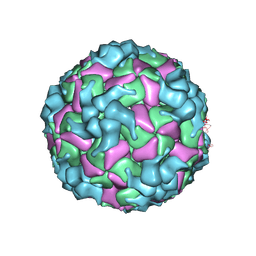 | | Cryo-EM structure of enterovirus A71 A-particle in complex with Fab h1A6.2 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Jiang, Y, Huang, Y, Zhu, R, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-11-29 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Cryo-EM structure of enterovirus A71 A-particle in complex with Fab h1A6.2
To Be Published
|
|
8X99
 
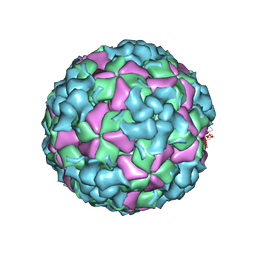 | | Cryo-EM structure of coxsackievirus A16 A-particle in complex with Fab h1A6.2 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Jiang, Y, Huang, Y, Zhu, R, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-11-29 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Cryo-EM structure of coxsackievirus A16 A-particle in complex with Fab h1A6.2
To Be Published
|
|
8X95
 
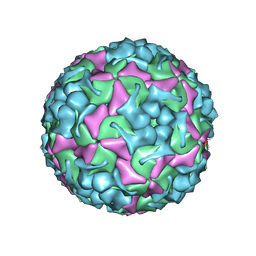 | | Cryo-EM structure of enterovirus A71 mature virion in complex with Fab h1A6.2 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Jiang, Y, Huang, Y, Zhu, R, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-11-29 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Cryo-EM structure of enterovirus A71 mature virion in complex with Fab h1A6.2
To Be Published
|
|
8X97
 
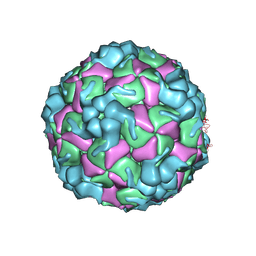 | | Cryo-EM structure of enterovirus A71 empty particle in complex with Fab h1A6.2 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Jiang, Y, Huang, Y, Zhu, R, Zheng, Q, Li, S. | | Deposit date: | 2023-11-29 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Cryo-EM structure of enterovirus A71 empty particle in complex with Fab h1A6.2
To Be Published
|
|
8X9A
 
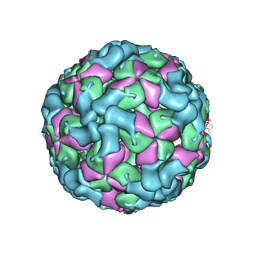 | | Cryo-EM structure of coxsackievirus A16 empty particle in complex with Fab h1A6.2 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Jiang, Y, Huang, Y, Zhu, R, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-11-29 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Cryo-EM structure of coxsackievirus A16 empty particle in complex with Fab h1A6.2
To Be Published
|
|
2L9R
 
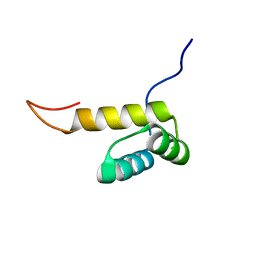 | | Solution NMR Structure of Homeobox domain of Homeobox protein Nkx-3.1 from homo sapiens, Northeast Structural Genomics Consortium Target HR6470A | | Descriptor: | Homeobox protein Nkx-3.1 | | Authors: | Liu, G, Xiao, R, Lee, H.-W, Hamilton, K, Ciccosanti, C, Wang, H.B, Acton, T.B, Everett, J.K, Huang, Y.J, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-02-22 | | Release date: | 2011-04-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Homeobox domain of Homeobox protein Nkx-3.1 from homo sapiens, Northeast Structural Genomics Consortium Target HR6470A
To be Published
|
|
2LA6
 
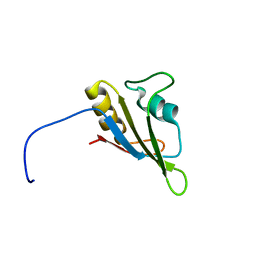 | | Solution NMR Structure of RRM domain of RNA-binding protein FUS from homo sapiens, Northeast Structural Genomics Consortium Target HR6430A | | Descriptor: | RNA-binding protein FUS | | Authors: | Liu, G, Xiao, R, Janjua, H, Ciccosanti, C, Wang, H, Lee, H, Acton, T.B, Everett, J.K, Huang, Y.J, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-03-04 | | Release date: | 2011-05-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Northeast Structural Genomics Consortium Target HR6430A
To be Published
|
|
7CH6
 
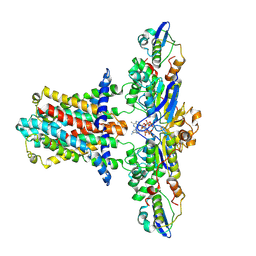 | | Cryo-EM structure of E.coli MlaFEB with AMPPNP | | Descriptor: | Lipid asymmetry maintenance ABC transporter permease subunit MlaE, Lipid asymmetry maintenance protein MlaB, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Zhou, C, Shi, H, Zhang, M, Huang, Y. | | Deposit date: | 2020-07-05 | | Release date: | 2021-08-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Insight into Phospholipid Transport by the MlaFEBD Complex from P. aeruginosa.
J.Mol.Biol., 433, 2021
|
|
7CH8
 
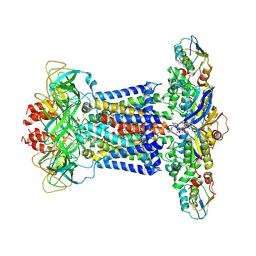 | | Cryo-EM structure of P.aeruginosa MlaFEBD with ADP-V | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, ADP METAVANADATE, MAGNESIUM ION, ... | | Authors: | Zhou, C, Shi, H, Zhang, M, Huang, Y. | | Deposit date: | 2020-07-05 | | Release date: | 2021-10-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Insight into Phospholipid Transport by the MlaFEBD Complex from P. aeruginosa.
J.Mol.Biol., 433, 2021
|
|
7CH9
 
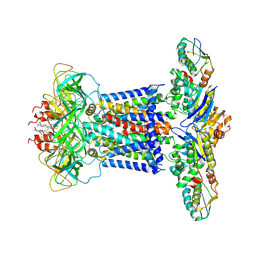 | | Cryo-EM structure of P.aeruginosa MlaFEBD | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, MlaD domain-containing protein, Probable ATP-binding component of ABC transporter, ... | | Authors: | Zhou, C, Shi, H, Zhang, M, Huang, Y. | | Deposit date: | 2020-07-05 | | Release date: | 2021-10-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Insight into Phospholipid Transport by the MlaFEBD Complex from P. aeruginosa.
J.Mol.Biol., 433, 2021
|
|
6L0I
 
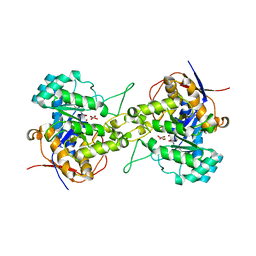 | | Crystal structure of dihydroorotase in complex with malate at pH6.5 from Saccharomyces cerevisiae | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Dihydroorotase, ZINC ION | | Authors: | Guan, H.H, Huang, Y.H, Huang, C.Y, Chen, C.J. | | Deposit date: | 2019-09-26 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the interaction modes of dihydroorotase with the anticancer drugs 5-fluorouracil and 5-aminouracil.
Biochem.Biophys.Res.Commun., 551, 2021
|
|
6L0F
 
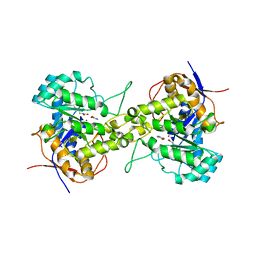 | | Crystal structure of dihydroorotase in complex with 5-Aminouracil from Saccharomyces cerevisiae | | Descriptor: | 5-AMINO-1H-PYRIMIDINE-2,4-DIONE, Dihydroorotase, ZINC ION | | Authors: | Guan, H.H, Huang, Y.H, Huang, C.Y, Chen, C.J. | | Deposit date: | 2019-09-26 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | Structural basis for the interaction modes of dihydroorotase with the anticancer drugs 5-fluorouracil and 5-aminouracil.
Biochem.Biophys.Res.Commun., 551, 2021
|
|
7CH7
 
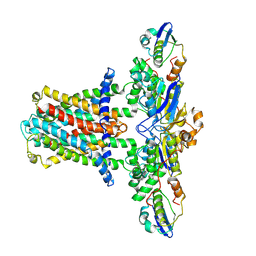 | | Cryo-EM structure of E.coli MlaFEB | | Descriptor: | Lipid asymmetry maintenance ABC transporter permease subunit MlaE, Lipid asymmetry maintenance protein MlaB, Phospholipid ABC transporter ATP-binding protein MlaF | | Authors: | Zhou, C, Shi, H, Huang, Y. | | Deposit date: | 2020-07-05 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Insight into Phospholipid Transport by the MlaFEBD Complex from P. aeruginosa.
J.Mol.Biol., 433, 2021
|
|
7CHA
 
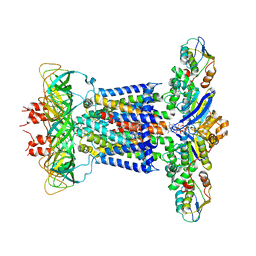 | | Cryo-EM structure of P.aeruginosa MlaFEBD with AMPPNP | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, MlaD domain-containing protein, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Zhou, C, Shi, H, Zhang, M, Huang, Y. | | Deposit date: | 2020-07-05 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Insight into Phospholipid Transport by the MlaFEBD Complex from P. aeruginosa.
J.Mol.Biol., 433, 2021
|
|
2M5O
 
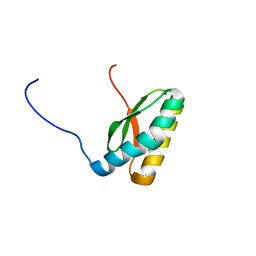 | | Solution NMR Structure CTD domain of NFU1 Iron-Sulfur Cluster Scaffold Homolog from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR2876C | | Descriptor: | NFU1 iron-sulfur cluster scaffold homolog, mitochondrial | | Authors: | Liu, G, Xiao, R, Janjua, H, Hamilton, K, Shastry, R, Kohan, E, Acton, T.B, Everett, J.K, Pederson, K, Huang, Y.J, Montelione, G.T, Northeast Structural Genomics Consortium (NESG), Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2013-03-01 | | Release date: | 2013-05-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure CTD domain of NFU1 Iron-Sulfur Cluster Scaffold Homolog from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR2876C
To be Published
|
|
6L0K
 
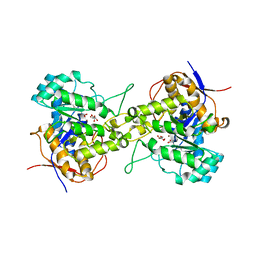 | | Crystal structure of dihydroorotase in complex with malate at pH9 from Saccharomyces cerevisiae | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Dihydroorotase, ZINC ION | | Authors: | Guan, H.H, Huang, Y.H, Huang, C.Y, Chen, C.J. | | Deposit date: | 2019-09-26 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for the interaction modes of dihydroorotase with the anticancer drugs 5-fluorouracil and 5-aminouracil.
Biochem.Biophys.Res.Commun., 551, 2021
|
|
6L0J
 
 | | Crystal structure of Dihydroorotase in complex with malate at pH7.5 from Saccharomyces cerevisiae | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Dihydroorotase, ZINC ION | | Authors: | Guan, H.H, Huang, Y.H, Huang, C.Y, Chen, C.J. | | Deposit date: | 2019-09-26 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.933 Å) | | Cite: | Structural Analysis of Saccharomyces cerevisiae Dihydroorotase Reveals Molecular Insights into the Tetramerization Mechanism
Molecules, 2021
|
|
