9O55
 
 | | Structure of a synthetic antibody (RM010) in complex with a class I MHC presenting a hapten-peptide conjugate | | Descriptor: | GTPase KRas, N-terminally processed, MHC class I antigen, ... | | Authors: | Hu, Z, Rajak, E, Maso, L, Koide, S. | | Deposit date: | 2025-04-09 | | Release date: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Generation of actionable, cancer-specific neoantigens from KRAS(G12C) with adagrasib
To Be Published
|
|
6PXZ
 
 | |
6PY0
 
 | |
6INU
 
 | |
6INN
 
 | |
6INV
 
 | |
8EU3
 
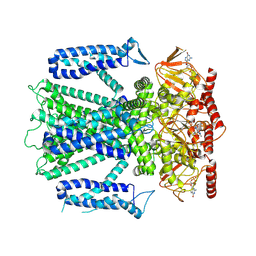 | | Cryo-EM structure of cGMP bound human CNGA3/CNGB3 channel in GDN, transition state 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYCLIC GUANOSINE MONOPHOSPHATE, Cyclic nucleotide-gated cation channel alpha-3, ... | | Authors: | Hu, Z, Zheng, X, Yang, J. | | Deposit date: | 2022-10-18 | | Release date: | 2023-08-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Conformational trajectory of allosteric gating of the human cone photoreceptor cyclic nucleotide-gated channel.
Nat Commun, 14, 2023
|
|
8ETP
 
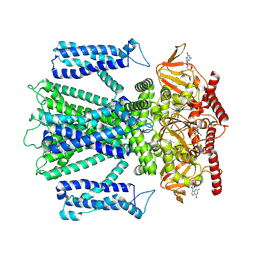 | | Cryo-EM structure of cGMP bound closed state of human CNGA3/CNGB3 channel in GDN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYCLIC GUANOSINE MONOPHOSPHATE, Cyclic nucleotide-gated cation channel alpha-3, ... | | Authors: | Hu, Z, Zheng, X, Yang, J. | | Deposit date: | 2022-10-17 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Conformational trajectory of allosteric gating of the human cone photoreceptor cyclic nucleotide-gated channel.
Nat Commun, 14, 2023
|
|
8EUC
 
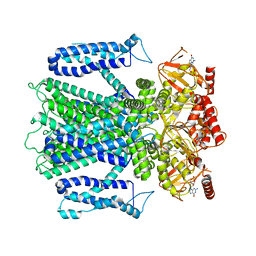 | | Cryo-EM structure of cGMP bound human CNGA3/CNGB3 channel in GDN, transition state 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYCLIC GUANOSINE MONOPHOSPHATE, Cyclic nucleotide-gated cation channel alpha-3, ... | | Authors: | Hu, Z, Zheng, X, Yang, J. | | Deposit date: | 2022-10-18 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Conformational trajectory of allosteric gating of the human cone photoreceptor cyclic nucleotide-gated channel.
Nat Commun, 14, 2023
|
|
8EVA
 
 | |
8EVB
 
 | |
8EV9
 
 | |
8EVC
 
 | |
8EV8
 
 | |
8XPU
 
 | | Overall structure of the LAT1-4F2hc bound with JPH203 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 5 of Amino acid transporter heavy chain SLC3A2, Large neutral amino acids transporter small subunit 1, ... | | Authors: | Hu, Z, Yan, R. | | Deposit date: | 2024-01-04 | | Release date: | 2024-07-17 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for the inhibition mechanism of LAT1-4F2hc complex by JPH203.
Cell Discov, 10, 2024
|
|
9EH6
 
 | | Crystal Structure of AroC | | Descriptor: | Alpha/beta hydrolase fold-5 domain-containing protein | | Authors: | Hu, Z, Klupt, K, Zechel, D, Jia, Z, Howe, G. | | Deposit date: | 2024-11-22 | | Release date: | 2025-02-26 | | Last modified: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mining Thermophile Genomes for New PETases with Exceptional Thermostabilities Using Sequence Similarity Networks.
Chembiochem, 26, 2025
|
|
6INO
 
 | |
6IOE
 
 | |
8WBY
 
 | | Cryo-EM structure of ACE2-B0AT1 complex with JX98 | | Descriptor: | 2-(4-chloranyl-3,5-dimethyl-phenoxy)-~{N}-propan-2-yl-ethanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, R, Hu, Z, Dai, L. | | Deposit date: | 2023-09-10 | | Release date: | 2024-09-11 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Molecular basis of inhibition of the amino acid transporter B 0 AT1 (SLC6A19).
Nat Commun, 15, 2024
|
|
6OC9
 
 | | S8 phosphorylated beta amyloid 40 fibrils | | Descriptor: | Amyloid-beta precursor protein, PHOSPHONATE | | Authors: | Qiang, W, Hu, Z.W. | | Deposit date: | 2019-03-22 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Molecular structure of an N-terminal phosphorylated beta-amyloid fibril.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8WBZ
 
 | | Cryo-EM structure of ACE2-B0AT1 complex with JX225 | | Descriptor: | 2-(4-bromanyl-3-methyl-phenoxy)-~{N}-propyl-ethanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, R, Hu, Z, Dai, L. | | Deposit date: | 2023-09-10 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis of inhibition of the amino acid transporter B 0 AT1 (SLC6A19).
Nat Commun, 15, 2024
|
|
7KOG
 
 | | Lethocerus Myosin II complete coiled-coil domain resolved in its native environment | | Descriptor: | Myosin heavy chain isoform Mhc_X1 | | Authors: | Rahmani, H, Hu, Z, Daneshparvar, N, Taylor, D, Taylor, K.A. | | Deposit date: | 2020-11-09 | | Release date: | 2021-03-24 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | The myosin II coiled-coil domain atomic structure in its native environment.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5J3I
 
 | | NMR solution structure of [Sp, Sp]-PT dsDNA | | Descriptor: | DNA (5'-D(*CP*GP*(SSG)P*CP*CP*GP*CP*CP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*GP*CP*GP*(SSG)P*CP*CP*G)-3') | | Authors: | Lan, W, Hu, Z, Cao, C. | | Deposit date: | 2016-03-30 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural investigation into physiological DNA phosphorothioate modification
Sci Rep, 6, 2016
|
|
5J3F
 
 | | NMR solution structure of [Rp, Rp]-PT dsDNA | | Descriptor: | DNA (5'-D(*CP*GP*(RSG)P*CP*CP*GP*CP*CP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*GP*CP*GP*(RSG)P*CP*CP*G)-3') | | Authors: | Lan, W, Hu, Z, Cao, C. | | Deposit date: | 2016-03-30 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural investigation into physiological DNA phosphorothioate modification
Sci Rep, 6, 2016
|
|
7N16
 
 | | Structure of TAX-4_R421W apo closed state | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cyclic nucleotide-gated cation channel, SODIUM ION | | Authors: | Zheng, X, Li, H, Hu, Z, Su, D, Yang, J. | | Deposit date: | 2021-05-27 | | Release date: | 2022-03-16 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional characterization of an achromatopsia-associated mutation in a phototransduction channel.
Commun Biol, 5, 2022
|
|
