5BRQ
 
 | |
5BRP
 
 | |
4S05
 
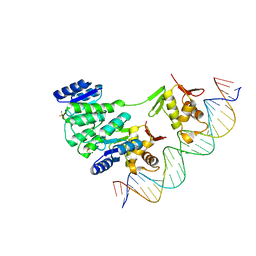 | | Crystal structure of Klebsiella pneumoniae PmrA in complex with PmrA box DNA | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, DNA (26-MER), DNA-binding transcriptional regulator BasR, ... | | Authors: | Hsiao, C.D, Weng, T.H, Li, Y.C. | | Deposit date: | 2014-12-30 | | Release date: | 2015-11-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure and dynamics of polymyxin-resistance-associated response regulator PmrA in complex with promoter DNA.
Nat Commun, 6, 2015
|
|
4S04
 
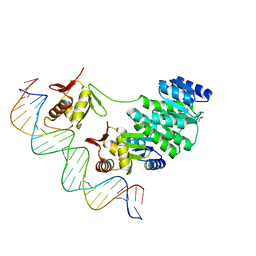 | | Crystal structure of Klebsiella pneumoniae PmrA in complex with PmrA box DNA | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, DNA (25-MER), DNA-binding transcriptional regulator BasR, ... | | Authors: | Hsiao, C.D, Weng, T.H, Li, Y.C. | | Deposit date: | 2014-12-30 | | Release date: | 2015-11-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and dynamics of polymyxin-resistance-associated response regulator PmrA in complex with promoter DNA.
Nat Commun, 6, 2015
|
|
1GGG
 
 | |
1E9L
 
 | |
8YK9
 
 | | Structure of Saccharolobus solfataricus SegC (SSO0033) protein, ATP soak | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SegC | | Authors: | Yen, C.Y, Lin, M.G, Sun, Y.J, Hsiao, C.D. | | Deposit date: | 2024-03-04 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Unraveling the structure and function of a novel SegC protein interacting with the SegAB chromosome segregation complex in Archaea.
Nucleic Acids Res., 52, 2024
|
|
1GSU
 
 | | AN AVIAN CLASS-MU GLUTATHIONE S-TRANSFERASE, CGSTM1-1 AT 1.94 ANGSTROM RESOLUTION | | Descriptor: | CLASS-MU GLUTATHIONE S-TRANSFERASE, S-HEXYLGLUTATHIONE | | Authors: | Sun, Y.-J, Kuan, C, Tam, M.F, Hsiao, C.-D. | | Deposit date: | 1997-09-02 | | Release date: | 1998-03-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The three-dimensional structure of an avian class-mu glutathione S-transferase, cGSTM1-1 at 1.94 A resolution.
J.Mol.Biol., 278, 1998
|
|
3ZQC
 
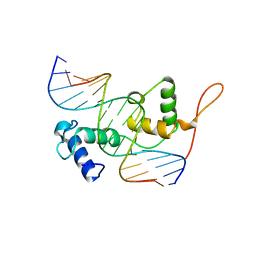 | | Structure of the Trichomonas vaginalis Myb3 DNA-binding domain bound to a promoter sequence reveals a unique C-terminal beta-hairpin conformation | | Descriptor: | MRE-1, MYB3 | | Authors: | Wei, S.-Y, Lou, Y.-C, Tsai, J.-Y, Hsu, H.-M, Tai, J.-H, Hsiao, C.-D, Chen, C. | | Deposit date: | 2011-06-09 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the Trichomonas Vaginalis Myb3 DNA-Binding Domain Bound to a Promoter Sequence Reveals a Unique C-Terminal Beta-Hairpin Conformation.
Nucleic Acids Res., 40, 2012
|
|
5WTN
 
 | |
8WQN
 
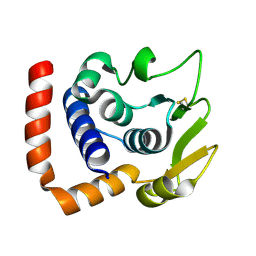 | |
8WQ8
 
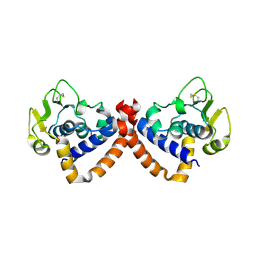 | |
2PGI
 
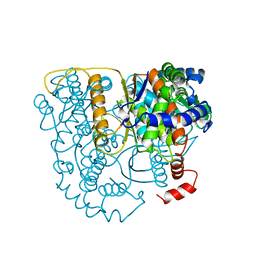 | | THE CRYSTAL STRUCTURE OF PHOSPHOGLUCOSE ISOMERASE-AN ENZYME WITH AUTOCRINE MOTILITY FACTOR ACTIVITY IN TUMOR CELLS | | Descriptor: | PHOSPHOGLUCOSE ISOMERASE | | Authors: | Sun, Y.-J, Chou, C.-C, Chen, W.-S, Meng, M, Hsiao, C.-D. | | Deposit date: | 1998-10-27 | | Release date: | 1999-06-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of a multifunctional protein: phosphoglucose isomerase/autocrine motility factor/neuroleukin.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1KXI
 
 | | STRUCTURE OF CYTOTOXIN HOMOLOG PRECURSOR | | Descriptor: | CARDIOTOXIN V | | Authors: | Sun, Y.-J, Wu, W.-G, Chiang, C.-M, Hsin, A.-Y, Hsiao, C.-D. | | Deposit date: | 1996-08-29 | | Release date: | 1997-04-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure of cardiotoxin V from Taiwan cobra venom: pH-dependent conformational change and a novel membrane-binding motif identified in the three-finger loops of P-type cardiotoxin.
Biochemistry, 36, 1997
|
|
1NY3
 
 | | Crystal structure of ADP bound to MAP KAP kinase 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAP kinase-activated protein kinase 2 | | Authors: | Underwood, K.W, Parris, K.D, Federico, E, Mosyak, L, Shane, T, Taylor, M, Svenson, K, Liu, Y, Hsiao, C.L, Wolfrom, S, Maguire, M, Malakian, K, Telliez, J.B, Lin, L.L, Kriz, R.W, Seehra, J, Somers, W.S, Stahl, M.L. | | Deposit date: | 2003-02-11 | | Release date: | 2003-10-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Catalytically active MAP KAP kinase 2 structures in complex with staurosporine and ADP reveal differences with the autoinhibited enzyme
Structure, 11, 2003
|
|
1C7R
 
 | | THE CRYSTAL STRUCTURE OF PHOSPHOGLUCOSE ISOMERASE/AUTOCRINE MOTILITY FACTOR/NEUROLEUKIN COMPLEXED WITH ITS CARBOHYDRATE PHOSPHATE INHIBITORS AND ITS SUBSTRATE RECOGNITION MECHANISM | | Descriptor: | 5-PHOSPHOARABINONIC ACID, PHOSPHOGLUCOSE ISOMERASE | | Authors: | Chou, C.-C, Meng, M, Sun, Y.-J, Hsiao, C.-D. | | Deposit date: | 2000-03-02 | | Release date: | 2000-09-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of phosphoglucose isomerase/autocrine motility factor/neuroleukin complexed with its carbohydrate phosphate inhibitors suggests its substrate/receptor recognition.
J.Biol.Chem., 275, 2000
|
|
1C7Q
 
 | | THE CRYSTAL STRUCTURE OF PHOSPHOGLUCOSE ISOMERASE/AUTOCRINE MOTILITY FACTOR/NEUROLEUKIN COMPLEXED WITH ITS CARBOHYDRATE PHOSPHATE INHIBITORS AND ITS SUBSTRATE RECOGNITION MECHANISM | | Descriptor: | N-BROMOACETYL-AMINOETHYL PHOSPHATE, PHOSPHOGLUCOSE ISOMERASE | | Authors: | Chou, C.-C, Meng, M, Sun, Y.-J, Hsiao, C.-D. | | Deposit date: | 2000-03-02 | | Release date: | 2000-09-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of phosphoglucose isomerase/autocrine motility factor/neuroleukin complexed with its carbohydrate phosphate inhibitors suggests its substrate/receptor recognition.
J.Biol.Chem., 275, 2000
|
|
6JSX
 
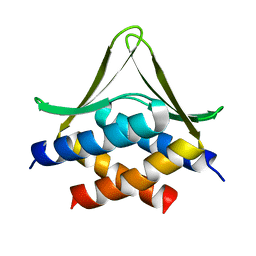 | |
1B0Z
 
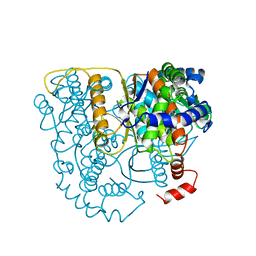 | | The crystal structure of phosphoglucose isomerase-an enzyme with autocrine motility factor activity in tumor cells | | Descriptor: | PROTEIN (PHOSPHOGLUCOSE ISOMERASE) | | Authors: | Sun, Y.-J, Chou, C.-C, Chen, W.-S, Meng, M, Hsiao, C.-D. | | Deposit date: | 1998-11-15 | | Release date: | 1999-11-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of phosphoglucose isomerase/autocrine motility factor/neuroleukin complexed with its carbohydrate phosphate inhibitors suggests its substrate/receptor recognition
J.Biol.Chem., 275, 2000
|
|
1NXK
 
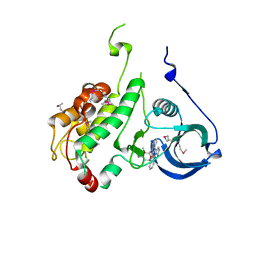 | | Crystal structure of staurosporine bound to MAP KAP kinase 2 | | Descriptor: | MAP kinase-activated protein kinase 2, STAUROSPORINE, SULFATE ION | | Authors: | Underwood, K.W, Parris, K.D, Federico, E, Mosyak, L, Czerwinski, R.M, Shane, T, Taylor, M, Svenson, K, Liu, Y, Hsiao, C.L, Wolfrom, S, Malakian, K, Telliez, J.B, Lin, L.L, Kriz, R.W, Seehra, J, Somers, W.S, Stahl, M.L. | | Deposit date: | 2003-02-10 | | Release date: | 2003-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Catalytically active MAP KAP kinase 2 structures in complex with staurosporine and ADP reveal differences with the autoinhibited enzyme
Structure, 11, 2003
|
|
1WDN
 
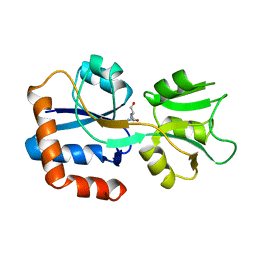 | | GLUTAMINE-BINDING PROTEIN | | Descriptor: | GLUTAMINE, GLUTAMINE BINDING PROTEIN | | Authors: | Sun, Y.-J, Rose, J, Wang, B.-C, Hsiao, C.-D. | | Deposit date: | 1997-05-17 | | Release date: | 1998-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The structure of glutamine-binding protein complexed with glutamine at 1.94 A resolution: comparisons with other amino acid binding proteins.
J.Mol.Biol., 278, 1998
|
|
1C72
 
 | | TYR115, GLN165 AND TRP209 CONTRIBUTE TO THE 1,2-EPOXY-3-(P-NITROPHENOXY)PROPANE CONJUGATING ACTIVITIES OF GLUTATHIONE S-TRANSFERASE CGSTM1-1 | | Descriptor: | 1-HYDROXY-2-S-GLUTATHIONYL-3-PARA-NITROPHENOXY-PROPANE, PROTEIN (GLUTATHIONE S-TRANSFERASE) | | Authors: | Chern, M.K, Wu, T.C, Hsieh, C.H, Chou, C.C, Liu, L.F, Kuan, I.C, Yeh, Y.H, Hsiao, C.D, Tam, M.F. | | Deposit date: | 2000-02-02 | | Release date: | 2000-08-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Tyr115, gln165 and trp209 contribute to the 1, 2-epoxy-3-(p-nitrophenoxy)propane-conjugating activity of glutathione S-transferase cGSTM1-1.
J.Mol.Biol., 300, 2000
|
|
2J3E
 
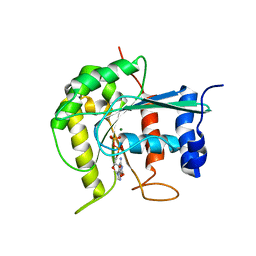 | | Dimerization is important for the GTPase activity of chloroplast translocon components atToc33 and psToc159 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, T7I23.11 PROTEIN | | Authors: | Yeh, Y.-H, Kesavulu, M.M, Wu, S.-Z, Li, H.-M, Sun, Y.-J, Konozy, E.H, Hsiao, C.-D. | | Deposit date: | 2006-08-21 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Dimerization is Important for the Gtpase Activity of Chloroplast Translocon Components Attoc33 and Pstoc159.
J.Biol.Chem., 282, 2007
|
|
4CLC
 
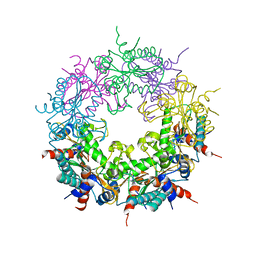 | | Crystal structure of Ybr137w protein | | Descriptor: | UPF0303 PROTEIN YBR137W | | Authors: | Yeh, Y.-H, Lin, T.-W, Lin, C.-Y, Hsiao, C.-D. | | Deposit date: | 2014-01-14 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Functional Characterization of Ybr137Wp Implicate its Involvement in the Targeting of Tail-Anchored Proteins to Membranes.
Mol.Cell.Biol., 34, 2014
|
|
2YIK
 
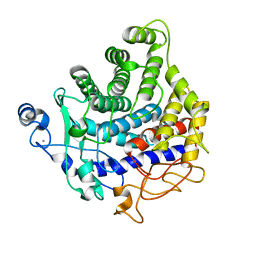 | | Catalytic domain of Clostridium thermocellum CelT | | Descriptor: | CALCIUM ION, ENDOGLUCANASE, ZINC ION | | Authors: | Tsai, J.-Y, Kesavulu, M.M, Hsiao, C.-D. | | Deposit date: | 2011-05-16 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the Catalytic Domain of the Clostridium Thermocellum Cellulase Celt
Acta Crystallogr.,Sect.D, 68, 2012
|
|
