3H73
 
 | |
3H72
 
 | |
7Y5F
 
 | | Crystal structure of CmnC in complex with L-homoarginine | | Descriptor: | CmnC, FE (III) ION, L(+)-TARTARIC ACID, ... | | Authors: | Hsiao, Y.H, Huang, S.J, Lin, E.C, Lee, Y.C, Chang, C.Y. | | Deposit date: | 2022-06-17 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal structure of the alpha-ketoglutarate-dependent non-heme iron oxygenase CmnC in capreomycin biosynthesis and its engineering to catalyze hydroxylation of the substrate enantiomer.
Front Chem, 10, 2022
|
|
7Y5I
 
 | | Crystal structure of CmnC in complex with L-homoarginine | | Descriptor: | ARGININE, CmnC, FE (III) ION, ... | | Authors: | Hsiao, Y.H, Huang, S.J, Lin, E.C, Lee, Y.C, Chang, C.Y. | | Deposit date: | 2022-06-17 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal structure of the alpha-ketoglutarate-dependent non-heme iron oxygenase CmnC in capreomycin biosynthesis and its engineering to catalyze hydroxylation of the substrate enantiomer.
Front Chem, 10, 2022
|
|
7Y5P
 
 | | Crystal structure of CmnC in complex with L-arginine and alpha-KG | | Descriptor: | 2-OXOGLUTARIC ACID, ARGININE, CmnC, ... | | Authors: | Hsiao, Y.H, Huang, S.J, Lin, E.C, Lee, Y.C, Chang, C.Y. | | Deposit date: | 2022-06-17 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the alpha-ketoglutarate-dependent non-heme iron oxygenase CmnC in capreomycin biosynthesis and its engineering to catalyze hydroxylation of the substrate enantiomer.
Front Chem, 10, 2022
|
|
3H71
 
 | |
2H4T
 
 | | Crystal structure of rat carnitine palmitoyltransferase II | | Descriptor: | Carnitine O-palmitoyltransferase II, mitochondrial, DODECANE | | Authors: | Hsiao, Y.S, Jogl, G, Esser, V, Tong, L. | | Deposit date: | 2006-05-25 | | Release date: | 2006-07-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of rat carnitine palmitoyltransferase II (CPT-II).
Biochem.Biophys.Res.Commun., 346, 2006
|
|
5DK5
 
 | | Crystal structure of CRN-4-MES complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cell death-related nuclease 4, ISOPROPYL ALCOHOL, ... | | Authors: | Hsiao, Y.-Y, Yuan, H.S. | | Deposit date: | 2015-09-03 | | Release date: | 2016-08-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of Inhibitors for the DEDDh Family of Exonucleases and a Unique Inhibition Mechanism by Crystal Structure Analysis of CRN-4 Bound with 2-Morpholin-4-ylethanesulfonate (MES)
J.Med.Chem., 59, 2016
|
|
1T7N
 
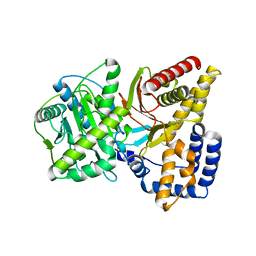 | |
1T7Q
 
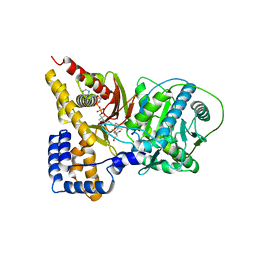 | | Crystal structure of the F565A mutant of murine carnitine acetyltransferase in complex with carnitine and CoA | | Descriptor: | 1,2-ETHANEDIOL, CARNITINE, COENZYME A, ... | | Authors: | Hsiao, Y.-S, Jogl, G, Tong, L. | | Deposit date: | 2004-05-10 | | Release date: | 2004-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and biochemical studies of the substrate selectivity of carnitine acetyltransferase
J.Biol.Chem., 279, 2004
|
|
1T7O
 
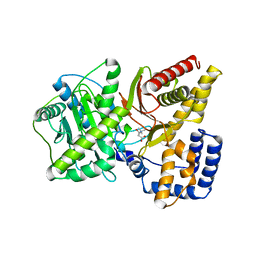 | |
5E2J
 
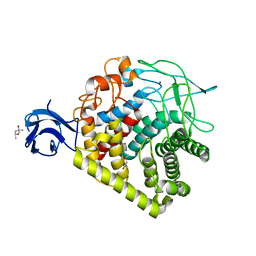 | | Crystal structure of single mutant thermostable endoglucanase (D468A) from Alicyclobacillus acidocaldarius | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Endoglucanase, ... | | Authors: | Hsiao, Y.Y, Wang, H.J, Tseng, C.P. | | Deposit date: | 2015-10-01 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Polarity Alteration of a Calcium Site Induces a Hydrophobic Interaction Network and Enhances Cel9A Endoglucanase Thermostability.
Appl.Environ.Microbiol., 82, 2016
|
|
2H3U
 
 | |
3H6J
 
 | |
2H3W
 
 | | Crystal structure of the S554A/M564G mutant of murine carnitine acetyltransferase in complex with hexanoylcarnitine and CoA | | Descriptor: | (R)-3-CARBOXY-2-(HEXANOYLOXY)-N,N,N-TRIMETHYLPROPAN-1-AMINIUM, COENZYME A, carnitine acetyltransferase | | Authors: | Hsiao, Y.S, Jogl, G, Tong, L. | | Deposit date: | 2006-05-23 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of murine carnitine acetyltransferase in ternary complexes with its substrates
J.Biol.Chem., 281, 2006
|
|
2H3P
 
 | | Crystal structure of murine carnitine acetyltransferase in complex with carnitine and acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, CARNITINE, COENZYME A, ... | | Authors: | Hsiao, Y.S, Jogl, G, Tong, L. | | Deposit date: | 2006-05-22 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of murine carnitine acetyltransferase in ternary complexes with its substrates
J.Biol.Chem., 281, 2006
|
|
3CG7
 
 | |
3CM5
 
 | |
3CM6
 
 | |
4KB1
 
 | |
4KAZ
 
 | | Crystal structure of RNase T in complex with a Y structured DNA | | Descriptor: | DNA (5'-D(*TP*TP*GP*GP*CP*CP*CP*TP*CP*TP*TP*TP*AP*GP*GP*GP*CP*CP*CP*C)-3'), MAGNESIUM ION, Ribonuclease T | | Authors: | Hsiao, Y.-Y, Yuan, H.S. | | Deposit date: | 2013-04-23 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into DNA repair by RNase T--an exonuclease processing 3' end of structured DNA in repair pathways.
Plos Biol., 12, 2014
|
|
4KB0
 
 | |
3NH0
 
 | |
3NGZ
 
 | |
3NH2
 
 | |
