5MBS
 
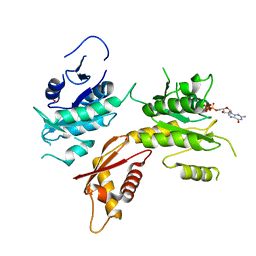 | |
1YR6
 
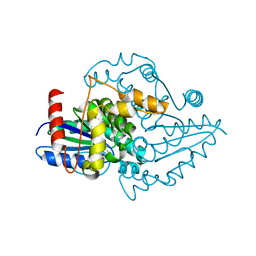 | |
1YR7
 
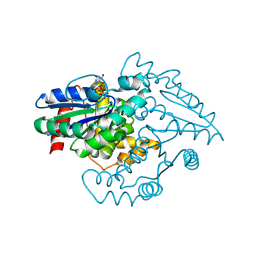 | | PAB0955 crystal structure : a GTPase in GTP-gamma-S bound form from Pyrococcus abyssi | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ATP(GTP)binding protein | | Authors: | Gras, S, Carpentier, P, Armengaud, J, Housset, D. | | Deposit date: | 2005-02-03 | | Release date: | 2006-02-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural insights into a new homodimeric self-activated GTPase family.
Embo Rep., 8, 2007
|
|
1YR8
 
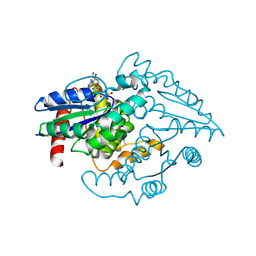 | | PAB0955 crystal structure : a GTPase in GTP bound form from Pyrococcus abyssi | | Descriptor: | ATP(GTP)binding protein, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Gras, S, Carpentier, P, Armengaud, J, Housset, D. | | Deposit date: | 2005-02-03 | | Release date: | 2006-02-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into a new homodimeric self-activated GTPase family.
Embo Rep., 8, 2007
|
|
1AHO
 
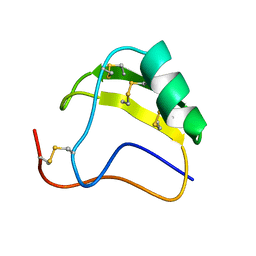 | | THE AB INITIO STRUCTURE DETERMINATION AND REFINEMENT OF A SCORPION PROTEIN TOXIN | | Descriptor: | TOXIN II | | Authors: | Smith, G.D, Blessing, R.H, Ealick, S.E, Fontecilla-Camps, J.C, Hauptman, H.A, Housset, D, Langs, D.A, Miller, R. | | Deposit date: | 1997-04-08 | | Release date: | 1997-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Ab initio structure determination and refinement of a scorpion protein toxin.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
1FO0
 
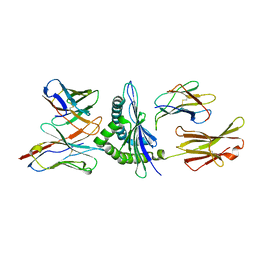 | | MURINE ALLOREACTIVE SCFV TCR-PEPTIDE-MHC CLASS I MOLECULE COMPLEX | | Descriptor: | NATURALLY PROCESSED OCTAPEPTIDE PBM1, PROTEIN (ALLOGENEIC H-2KB MHC CLASS I MOLECULE), PROTEIN (BETA-2 MICROGLOBULIN), ... | | Authors: | Reiser, J.B, Darnault, C, Guimezanes, A, Gregoire, C, Mosser, T, Schmitt-Verhulst, A.-M, Fontecilla-Camps, J.C, Malissen, B, Housset, D, Mazza, G. | | Deposit date: | 2000-08-24 | | Release date: | 2000-10-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a T cell receptor bound to an allogeneic MHC molecule.
Nat.Immunol., 1, 2000
|
|
3ECA
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI L-ASPARAGINASE, AN ENZYME USED IN CANCER THERAPY (ELSPAR) | | Descriptor: | ASPARTIC ACID, L-asparaginase 2 | | Authors: | Swain, A.L, Jaskolski, M, Housset, D, Rao, J.K.M, Wlodawer, A. | | Deposit date: | 1993-07-02 | | Release date: | 1993-10-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Escherichia coli L-asparaginase, an enzyme used in cancer therapy.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
1FDU
 
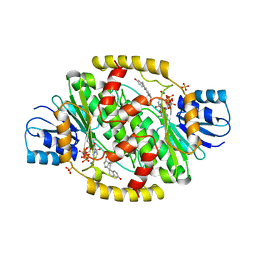 | | HUMAN 17-BETA-HYDROXYSTEROID-DEHYDROGENASE TYPE 1 MUTANT H221L COMPLEXED WITH ESTRADIOL AND NADP+ | | Descriptor: | 17-BETA-HYDROXYSTEROID DEHYDROGENASE, ESTRADIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Mazza, C, Breton, R, Housset, D, Fontecilla-Camps, J.-C. | | Deposit date: | 1998-01-14 | | Release date: | 1998-05-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Unusual charge stabilization of NADP+ in 17beta-hydroxysteroid dehydrogenase.
J.Biol.Chem., 273, 1998
|
|
1FDV
 
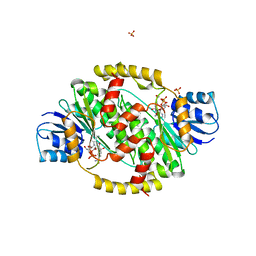 | | HUMAN 17-BETA-HYDROXYSTEROID-DEHYDROGENASE TYPE 1 MUTANT H221L COMPLEXED WITH NAD+ | | Descriptor: | 17-BETA-HYDROXYSTEROID DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Mazza, C, Breton, R, Housset, D, Fontecilla-Camps, J.-C. | | Deposit date: | 1998-01-15 | | Release date: | 1998-05-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Unusual charge stabilization of NADP+ in 17beta-hydroxysteroid dehydrogenase.
J.Biol.Chem., 273, 1998
|
|
1PTX
 
 | |
1FDW
 
 | | HUMAN 17-BETA-HYDROXYSTEROID-DEHYDROGENASE TYPE 1 MUTANT H221Q COMPLEXED WITH ESTRADIOL | | Descriptor: | 17-BETA-HYDROXYSTEROID DEHYDROGENASE, ESTRADIOL | | Authors: | Mazza, C, Breton, R, Housset, D, Fontecilla-Camps, J.-C. | | Deposit date: | 1998-01-16 | | Release date: | 1998-05-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Unusual charge stabilization of NADP+ in 17beta-hydroxysteroid dehydrogenase.
J.Biol.Chem., 273, 1998
|
|
4DCT
 
 | | Crystal Structure of B. subtilis EngA in complex with half-occupacy GDP | | Descriptor: | GTP-BINDING PROTEIN ENGA, GUANOSINE-5'-DIPHOSPHATE, SULFATE ION | | Authors: | Reiser, J.-B, Housset, D, Foucher, A.-E, Jault, J.-M. | | Deposit date: | 2012-01-18 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Potassium Acts as a GTPase-Activating Element on Each Nucleotide-Binding Domain of the Essential Bacillus subtilis EngA.
Plos One, 7, 2012
|
|
4DCU
 
 | | Crystal Structure of B. subtilis EngA in complex with GDP | | Descriptor: | GTP-BINDING PROTEIN ENGA, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Reiser, J.-B, Housset, D, Foucher, A.-E, Jault, J.-M. | | Deposit date: | 2012-01-18 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potassium Acts as a GTPase-Activating Element on Each Nucleotide-Binding Domain of the Essential Bacillus subtilis EngA.
Plos One, 7, 2012
|
|
3FT4
 
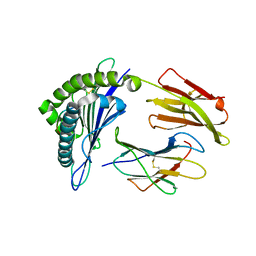 | | Crystal Structure of the minor histocompatibility peptide HA-1Arg in complex with HLA-A2 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Reiser, J.-B, Gras, S, Chouquet, A, Le Gorrec, M, Spierings, E, Goulmy, E, Housset, D. | | Deposit date: | 2009-01-12 | | Release date: | 2009-04-28 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Steric hindrance and fast dissociation explain the lack of immunogenicity of the minor histocompatibility HA-1Arg Null allele.
J.Immunol., 182, 2009
|
|
4DCS
 
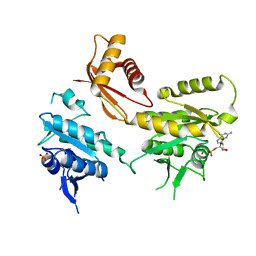 | | Crystal Structure of B. subtilis EngA in complex with sulfate ion and GDP | | Descriptor: | GTP-BINDING PROTEIN ENGA, GUANOSINE-5'-DIPHOSPHATE, SULFATE ION | | Authors: | Reiser, J.-B, Housset, D, Foucher, A.-E, Jault, J.-M. | | Deposit date: | 2012-01-18 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Potassium Acts as a GTPase-Activating Element on Each Nucleotide-Binding Domain of the Essential Bacillus subtilis EngA.
Plos One, 7, 2012
|
|
3FT2
 
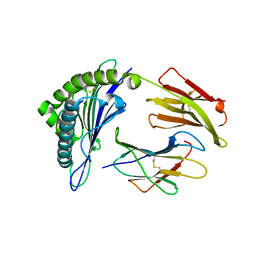 | | Crystal Structure of a citrulline peptide variant of the minor histocompatibility peptide HA-1 in complex with HLA-A2 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Reiser, J.-B, Gras, S, Chouquet, A, Le Gorrec, M, Spierings, E, Goulmy, E, Housset, D. | | Deposit date: | 2009-01-12 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Steric hindrance and fast dissociation explain the lack of immunogenicity of the minor histocompatibility HA-1Arg Null allele.
J.Immunol., 182, 2009
|
|
3FT3
 
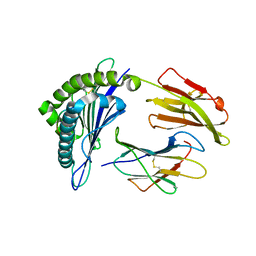 | | Crystal Structure of the minor histocompatibility peptide HA-1His in complex with HLA-A2 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Reiser, J.-B, Gras, S, Chouquet, A, Le Gorrec, M, Spierings, E, Goulmy, E, Housset, D. | | Deposit date: | 2009-01-12 | | Release date: | 2009-04-28 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Steric hindrance and fast dissociation explain the lack of immunogenicity of the minor histocompatibility HA-1Arg Null allele.
J.Immunol., 182, 2009
|
|
4DCV
 
 | | Crystal Structure of B. subtilis EngA in complex with GMPPCP | | Descriptor: | GTP-BINDING PROTEIN ENGA, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Reiser, J.-B, Housset, D, Foucher, A.-E, Jault, J.-M. | | Deposit date: | 2012-01-18 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Potassium Acts as a GTPase-Activating Element on Each Nucleotide-Binding Domain of the Essential Bacillus subtilis EngA.
Plos One, 7, 2012
|
|
1KJ3
 
 | | Mhc Class I H-2Kb molecule complexed with pKB1 peptide | | Descriptor: | BETA-2 MICROGLOBULIN, H-2KB MHC CLASS I MOLECULE ALPHA CHAIN, NATURALLY PROCESSED OCTAPEPTIDE PKB1 | | Authors: | Reiser, J.-B, Gregoire, C, Darnault, C, Mosser, T, Guimezanes, A, Schmitt-Verhulst, A.-M, Fontecilla-Camps, J.C, Mazza, G, Malissen, B, Housset, D. | | Deposit date: | 2001-12-04 | | Release date: | 2002-03-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A T cell receptor CDR3beta loop undergoes conformational changes of unprecedented magnitude upon binding to a peptide/MHC class I complex.
Immunity, 16, 2002
|
|
1A27
 
 | | HUMAN 17-BETA-HYDROXYSTEROID-DEHYDROGENASE TYPE 1 C-TERMINAL DELETION MUTANT COMPLEXED WITH ESTRADIOL AND NADP+ | | Descriptor: | 17-BETA-HYDROXYSTEROID-DEHYDROGENASE, ESTRADIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Mazza, C, Breton, R, Housset, D, Fontecilla-Camps, J.-C. | | Deposit date: | 1998-01-16 | | Release date: | 1998-05-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Human Type I 17Beta-Hydroxysteroid Dehydrogenase: Site Directed Mutagenesis and X-Ray Crystallography Structure-Function Analysis
Thesis, Universite Joseph Fourier, 1997
|
|
3GSN
 
 | | Crystal structure of the public RA14 TCR in complex with the HCMV dominant NLV/HLA-A2 epitope | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, HCMV pp65 fragment 495-503 (NLVPMVATV), ... | | Authors: | Gras, S, Saulquin, X, Reiser, J.-B, Debeaupuis, E, Echasserieau, K, Kissenpfennig, A, Legoux, F, Chouquet, A, Le Gorrec, M, Machillot, P, Neveu, B, Thielens, N, Malissen, B, Bonneville, M, Housset, D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-08-04 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural bases for the affinity-driven selection of a public TCR against a dominant human cytomegalovirus epitope.
J.Immunol., 183, 2009
|
|
3GSU
 
 | | Crystal structure of the binary complex between HLA-A2 and HCMV NLV-M5T peptide variant | | Descriptor: | Beta-2-microglobulin, HCMV pp65 fragment 495-503, variant M5T (NLVPTVATV), ... | | Authors: | Reiser, J.-B, Saulquin, X, Gras, S, Debeaupuis, E, Echasserieau, K, Kissenpfennig, A, Legoux, F, Chouquet, A, Le Gorrec, M, Machillot, P, Neveu, B, Thielens, N, Malissen, B, Bonneville, M, Housset, D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-08-04 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural bases for the affinity-driven selection of a public TCR against a dominant human cytomegalovirus epitope.
J.Immunol., 183, 2009
|
|
3GSX
 
 | | Crystal structure of the binary complex between HLA-A2 and HCMV NLV-T8V peptide variant | | Descriptor: | Beta-2-microglobulin, HCMV pp65 fragment 495-503, variant T8V (NLVPMVAVV), ... | | Authors: | Reiser, J.-B, Saulquin, X, Gras, S, Debeaupuis, E, Echasserieau, K, Kissenpfennig, A, Legoux, F, Chouquet, A, Le Gorrec, M, Machillot, P, Neveu, B, Thielens, N, Malissen, B, Bonneville, M, Housset, D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-08-04 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural bases for the affinity-driven selection of a public TCR against a dominant human cytomegalovirus epitope.
J.Immunol., 183, 2009
|
|
3GSR
 
 | | Crystal structure of the binary complex between HLA-A2 and HCMV NLV-M5V peptide variant | | Descriptor: | Beta-2-microglobulin, HCMV pp65 fragment 495-503, variant M5V (NLVPVVATV), ... | | Authors: | Reiser, J.-B, Saulquin, X, Gras, S, Debeaupuis, E, Echasserieau, K, Kissenpfennig, A, Legoux, F, Chouquet, A, Le Gorrec, M, Machillot, P, Neveu, B, Thielens, N, Malissen, B, Bonneville, M, Housset, D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-08-04 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural bases for the affinity-driven selection of a public TCR against a dominant human cytomegalovirus epitope.
J.Immunol., 183, 2009
|
|
3GSV
 
 | | Crystal structure of the binary complex between HLA-A2 and HCMV NLV-M5Q peptide variant | | Descriptor: | Beta-2-microglobulin, HCMV pp65 fragment 495-503, variant M5Q (NLVPQVATV), ... | | Authors: | Reiser, J.-B, Saulquin, X, Gras, S, Debeaupuis, E, Echasserieau, K, Kissenpfennig, A, Legoux, F, Chouquet, A, Le Gorrec, M, Machillot, P, Neveu, B, Thielens, N, Malissen, B, Bonneville, M, Housset, D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-08-04 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural bases for the affinity-driven selection of a public TCR against a dominant human cytomegalovirus epitope.
J.Immunol., 183, 2009
|
|
