8WKK
 
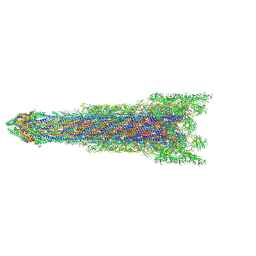 | | Cryo-EM structure of the whole rod with export apparatus and hook within the flagellar motor-hook complex in the CW state. | | Descriptor: | Flagellar M-ring protein, Flagellar basal body rod protein FlgB, Flagellar basal-body rod protein FlgC, ... | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2023-09-28 | | Release date: | 2024-09-04 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of the bacterial flagellar motor rotational switching.
Cell Res., 34, 2024
|
|
8HE4
 
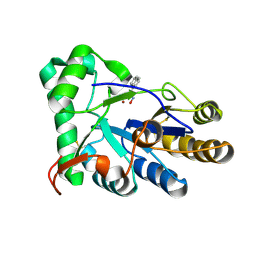 | | The structure of chitin deacetylase Pst_13661 from Puccinia striiformis f. sp. tritici | | Descriptor: | Chitin deacetylase, ZINC ION, ~{N}-oxidanylnaphthalene-1-carboxamide | | Authors: | Liu, L, Li, Y.C, Zhou, Y, Yang, Q. | | Deposit date: | 2022-11-07 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Inhibition of chitin deacetylases to attenuate plant fungal diseases.
Nat Commun, 14, 2023
|
|
8HE2
 
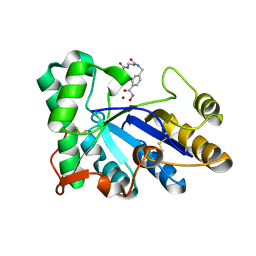 | | The structure of chitin deacetylase Pst_13661 from Puccinia striiformis f. sp. tritici | | Descriptor: | Chitin deacetylase, ZINC ION, tert-butyl N-[3-[[4-(oxidanylcarbamoyl)phenyl]methylamino]-3-oxidanylidene-propyl]carbamate | | Authors: | Liu, L, Li, Y.C, Zhou, Y, Yang, Q. | | Deposit date: | 2022-11-07 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Inhibition of chitin deacetylases to attenuate plant fungal diseases.
Nat Commun, 14, 2023
|
|
8HE1
 
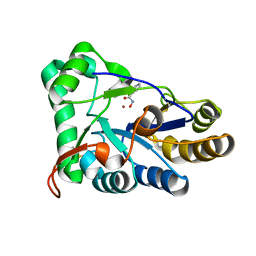 | | The structure of chitin deacetylase Pst_13661 from Puccinia striiformis f. sp. tritici | | Descriptor: | BENZHYDROXAMIC ACID, Chitin deacetylase, ZINC ION | | Authors: | Liu, L, Li, Y.C, Zhou, Y, Yang, Q. | | Deposit date: | 2022-11-07 | | Release date: | 2023-05-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Inhibition of chitin deacetylases to attenuate plant fungal diseases.
Nat Commun, 14, 2023
|
|
8HFA
 
 | |
8HF9
 
 | | The structure of chitin deacetylase Pst_13661 from Puccinia striiformis f. sp. tritici | | Descriptor: | Chitin deacetylase, ZINC ION | | Authors: | Liu, L, Li, Y.C, Zhou, Y, Yang, Q. | | Deposit date: | 2022-11-10 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Inhibition of chitin deacetylases to attenuate plant fungal diseases.
Nat Commun, 14, 2023
|
|
8YJT
 
 | | Cryo-EM structure of the flagellar C ring in the CCW state | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliM, ... | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2024-03-02 | | Release date: | 2024-09-04 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural basis of the bacterial flagellar motor rotational switching.
Cell Res., 34, 2024
|
|
8XP0
 
 | | Cryo-EM structure of the protomers of the C ring in the CCW state | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliM, ... | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2024-01-02 | | Release date: | 2024-09-04 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of the bacterial flagellar motor rotational switching.
Cell Res., 34, 2024
|
|
8XP1
 
 | | Cryo-EM structure of the protomers of the C ring in the CW state | | Descriptor: | Chemotaxis protein CheY, Flagellar M-ring protein, Flagellar motor switch protein FliG, ... | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2024-01-02 | | Release date: | 2024-09-04 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural basis of the bacterial flagellar motor rotational switching.
Cell Res., 34, 2024
|
|
8HTD
 
 | | Crystal structure of an effector from Chromobacterium violaceum in complex with ubiquitin | | Descriptor: | NAD(+)--protein-threonine ADP-ribosyltransferase, Ubiquitin | | Authors: | Tan, J, Wang, X, Zhou, Y, Zhu, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Molecular basis of threonine ADP-ribosylation of ubiquitin by bacterial ARTs.
Nat.Chem.Biol., 20, 2024
|
|
8HTE
 
 | | Crystal structure of an effector mutant in complex with ubiquitin | | Descriptor: | GLYCEROL, NAD(+)--protein-threonine ADP-ribosyltransferase, NICOTINAMIDE, ... | | Authors: | Tan, J, Wang, X, Zhou, Y, Zhu, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.307 Å) | | Cite: | Molecular basis of threonine ADP-ribosylation of ubiquitin by bacterial ARTs.
Nat.Chem.Biol., 20, 2024
|
|
8HTF
 
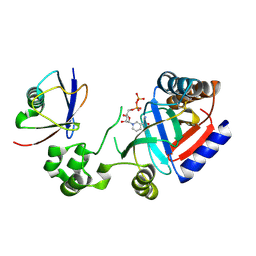 | | Crystal structure of an effector in complex with ubiquitin | | Descriptor: | NAD(+)--protein-threonine ADP-ribosyltransferase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Ubiquitin-40S ribosomal protein S27a (Fragment) | | Authors: | Tan, J, Wang, X, Zhou, Y, Zhu, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Molecular basis of threonine ADP-ribosylation of ubiquitin by bacterial ARTs.
Nat.Chem.Biol., 20, 2024
|
|
8HTC
 
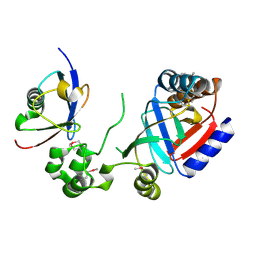 | | Crystal structure of a SeMet-labeled effector from Chromobacterium violaceum in complex with Ubiquitin | | Descriptor: | NAD(+)--protein-threonine ADP-ribosyltransferase, Ubiquitin-40S ribosomal protein S27a (Fragment) | | Authors: | Tan, J, Wang, X, Zhou, Y, Zhu, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis of threonine ADP-ribosylation of ubiquitin by bacterial ARTs.
Nat.Chem.Biol., 20, 2024
|
|
8WHT
 
 | | Cryo-EM structure of the LP ring within the flagellar motor-hook complex in the CW state | | Descriptor: | Flagellar L-ring protein, Flagellar P-ring protein | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2023-09-23 | | Release date: | 2024-09-04 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structural basis of the bacterial flagellar motor rotational switching.
Cell Res., 34, 2024
|
|
8WK3
 
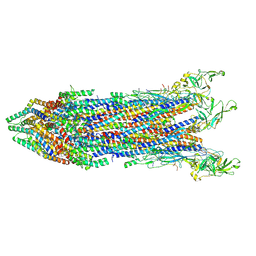 | | Cryo-EM structure of the proximal rod-export apparatus and FlgF within the motor-hook complex in the CW state | | Descriptor: | Flagellar M-ring protein, Flagellar basal body rod protein FlgB, Flagellar basal-body rod protein FlgC, ... | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2023-09-26 | | Release date: | 2024-09-04 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of the bacterial flagellar motor rotational switching.
Cell Res., 34, 2024
|
|
8WLE
 
 | | Cryo-EM structure of the LP ring within the flagellar motor-hook complex in the CCW state. | | Descriptor: | Flagellar L-ring protein, Flagellar P-ring protein | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2023-09-29 | | Release date: | 2024-09-04 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of the bacterial flagellar motor rotational switching.
Cell Res., 34, 2024
|
|
8WKQ
 
 | | Cryo-EM structure of the MS ring (C1) with export apparatus and proximal rod within the flagellar motor-hook complex in the CW state. | | Descriptor: | Flagellar M-ring protein, Flagellar basal body rod protein FlgB, Flagellar basal-body rod protein FlgC, ... | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2023-09-28 | | Release date: | 2024-09-04 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of the bacterial flagellar motor rotational switching.
Cell Res., 34, 2024
|
|
8J8Q
 
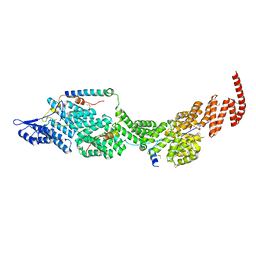 | | Structure of the four-component Paf1 complex from Saccharomyces eubayanus | | Descriptor: | CDC73-like protein, CTR9-like protein, PAF1-like protein, ... | | Authors: | Wang, Z, Qin, Y, Zhou, Y, Cao, Y. | | Deposit date: | 2023-05-02 | | Release date: | 2023-05-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural Basis of the Transcriptional Elongation Factor Paf1 Core Complex from Saccharomyces eubayanus .
Int J Mol Sci, 24, 2023
|
|
8J8P
 
 | | Structure of the four-component Paf1 complex from Saccharomyces eubayanus | | Descriptor: | CDC73-like protein, CTR9-like protein, PAF1-like protein, ... | | Authors: | Wang, Z, Qin, Y, Zhou, Y, Cao, Y. | | Deposit date: | 2023-05-02 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of the Transcriptional Elongation Factor Paf1 Core Complex from Saccharomyces eubayanus .
Int J Mol Sci, 24, 2023
|
|
2XNI
 
 | | Protein-ligand complex of a novel macrocyclic HCV NS3 protease inhibitor derived from amino cyclic boronates | | Descriptor: | (1-{[(10-tert-butyl-15,15-dimethyl-3,9,12-trioxo-6,7,9,10,11,12,14,15,16,17,18,19,23,23a-tetradecahydro-1H,5H-2,23:5,8-dimethano-4,13,2,8,11-benzodioxatriazacyclohenicosin-7(3H)-yl)carbonyl]amino}-3-hydroxypropyl)(trihydroxy)borate(1-), MAGNESIUM ION, NS3 PROTEASE, ... | | Authors: | Li, X, Zhang, Y.-K, Liu, Y, Ding, C.Z, Zhou, Y, Li, Q, Plattner, J.J, Baker, S.J, Zhang, S, Kazmierski, W.M, Wright, L.L, Smith, G.K, Grimes, R.M, Crosby, R.M, Creech, K.L, Carballo, L.H, Slater, M.J, Jarvest, R.L, Thommes, P, Hubbard, J.A, Convery, M.A, Nassau, P.M, McDowell, W, Skarzynski, T.J, Qian, X, Fan, D, Liao, L, Ni, Z.-J, Pennicott, L.E, Zou, W, Wright, J. | | Deposit date: | 2010-08-02 | | Release date: | 2011-08-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Novel Macrocyclic Hcv Ns3 Protease Inhibitors Derived from Alpha-Amino Cyclic Boronates.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
8YP3
 
 | | Crystal structure of UDP-N-acetylglucosamine pyrophosphorylase from Spodoptera frugiperda in complex with UDP-GlcNAc | | Descriptor: | MAGNESIUM ION, SULFATE ION, UDP-N-acetylglucosamine diphosphorylase, ... | | Authors: | Lu, Q, Liu, T, Zhou, Y, Yang, Q. | | Deposit date: | 2024-03-15 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure and Inhibition of Insect UDP- N -acetylglucosamine Pyrophosphorylase: A Key Enzyme in the Hexosamine Biosynthesis Pathway.
J.Agric.Food Chem., 72, 2024
|
|
7W0V
 
 | | C4'-SCF3-DT modifeid DNA-DNA duplex | | Descriptor: | DNA (5'-D(*CP*CP*AP*TP*(DSW)P*AP*TP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*TP*GP*G)-3') | | Authors: | Li, Q, Trajkovski, M, Fan, C, Chen, J, Zhou, Y, Lu, K, Li, H, Su, X, Xi, Z, Plavec, J, Zhou, C. | | Deposit date: | 2021-11-18 | | Release date: | 2022-11-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 4'-SCF 3 -Labeling Constitutes a Sensitive 19 F NMR Probe for Characterization of Interactions in the Minor Groove of DNA.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7VO5
 
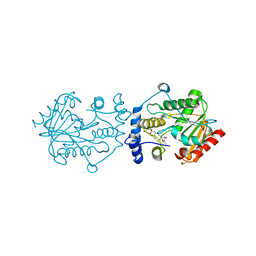 | | Pimaricin type I PKS thioesterase domain (holo Pim TE) | | Descriptor: | (1R,3S,5E,7S,11R,13E,15E,17E,19E,21R,23S,24R,25S)-11,24-dimethyl-1,3,7,21,25-pentakis(oxidanyl)-10,27-dioxabicyclo[21.3.1]heptacosa-5,13,15,17,19-pentaen-9-one, ScnS4 | | Authors: | Bai, L, Zhou, Y. | | Deposit date: | 2021-10-12 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Mechanistic Insights into Chain Release of the Polyene PKS Thioesterase Domain
Acs Catalysis, 12, 2022
|
|
7VO4
 
 | |
3IS1
 
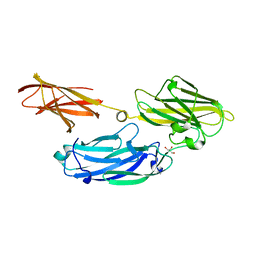 | | Crystal structure of functional region of UafA from Staphylococcus saprophyticus in C2 form at 2.45 angstrom resolution | | Descriptor: | GLYCEROL, Uro-adherence factor A | | Authors: | Tanaka, Y, Matsuoka, E, Shouji, Y, Kuroda, M, Tanaka, I, Yao, M. | | Deposit date: | 2009-08-24 | | Release date: | 2010-09-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of the functional region of Uro-adherence factor A from Staphylococcus saprophyticus reveals participation of the B domain in ligand binding
Protein Sci., 20, 2011
|
|
