8GAJ
 
 | | Crystal Structure of E. coli LptA in complex with Podisus maculiventris Thanatin | | Descriptor: | Lipopolysaccharide export system protein LptA, Thanatin | | Authors: | Huynh, K, Kibrom, A, Donald, B, Zhou, P. | | Deposit date: | 2023-02-23 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discovery, characterization, and redesign of potent antimicrobial thanatin orthologs from Chinavia ubica and Murgantia histrionica targeting E. coli LptA.
J Struct Biol X, 8, 2023
|
|
8GAL
 
 | | Crystal Structure of the E. coli LptA in complex with Murgantia histrionica Thanatin | | Descriptor: | Lipopolysaccharide export system protein LptA, Thanatin | | Authors: | Kibrom, A, Huynh, K, Donald, B, Zhou, P. | | Deposit date: | 2023-02-23 | | Release date: | 2023-07-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery, characterization, and redesign of potent antimicrobial thanatin orthologs from Chinavia ubica and Murgantia histrionica targeting E. coli LptA.
J Struct Biol X, 8, 2023
|
|
8GAK
 
 | | Crystal Structure of E. coli LptA in complex with Chinavia Ubica Thanatin | | Descriptor: | Lipopolysaccharide export system protein LptA, Thanatin | | Authors: | Huynh, K, Kibrom, A, Donald, B, Zhou, P. | | Deposit date: | 2023-02-23 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery, characterization, and redesign of potent antimicrobial thanatin orthologs from Chinavia ubica and Murgantia histrionica targeting E. coli LptA.
J Struct Biol X, 8, 2023
|
|
2MUR
 
 | |
1XXE
 
 | | RDC refined solution structure of the AaLpxC/TU-514 complex | | Descriptor: | 1,5-ANHYDRO-2-C-(CARBOXYMETHYL-N-HYDROXYAMIDE)-2-DEOXY-3-O-MYRISTOYL-D-GLUCITOL, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Coggins, B.E, McClerren, A.L, Jiang, L, Li, X, Rudolph, J, Hindsgaul, O, Raetz, C.R.H, Zhou, P. | | Deposit date: | 2004-11-04 | | Release date: | 2004-11-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined Solution Structure of the LpxC-TU-514 Complex and pK(a) Analysis of an Active Site Histidine: Insights into the Mechanism and Inhibitor Design
Biochemistry, 44, 2005
|
|
2MUQ
 
 | | Solution Structure of the Human FAAP20 UBZ | | Descriptor: | Fanconi anemia-associated protein of 20 kDa, ZINC ION | | Authors: | Wojtaszek, J.L, Wang, S, Zhou, P. | | Deposit date: | 2014-09-16 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ubiquitin recognition by FAAP20 expands the complex interface beyond the canonical UBZ domain.
Nucleic Acids Res., 42, 2014
|
|
2B5M
 
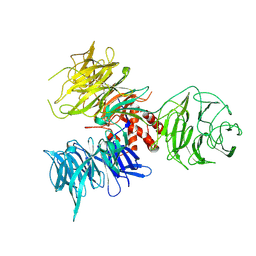 | | Crystal Structure of DDB1 | | Descriptor: | damage-specific DNA binding protein 1 | | Authors: | Li, T, Chen, X, Garbutt, K.C, Zhou, P, Zheng, N. | | Deposit date: | 2005-09-28 | | Release date: | 2006-02-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Structure of DDB1 in complex with a paramyxovirus V protein: viral hijack of a propeller cluster in ubiquitin ligase.
Cell(Cambridge,Mass.), 124, 2006
|
|
2B5N
 
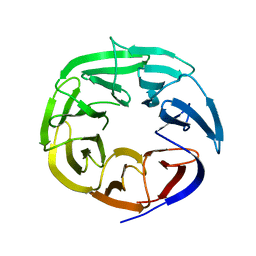 | | Crystal Structure of the DDB1 BPB Domain | | Descriptor: | ISOPROPYL ALCOHOL, damage-specific DNA binding protein 1 | | Authors: | Li, T, Chen, X, Garbutt, K.C, Zhou, P, Zheng, N. | | Deposit date: | 2005-09-28 | | Release date: | 2006-02-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of DDB1 in complex with a paramyxovirus V protein: viral hijack of a propeller cluster in ubiquitin ligase.
Cell(Cambridge,Mass.), 124, 2006
|
|
3WNV
 
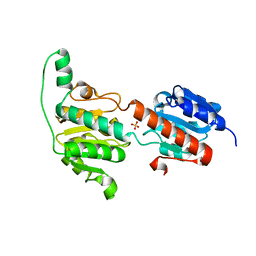 | | Crystal structure of a glyoxylate reductase from Paecilomyes thermophila | | Descriptor: | SULFATE ION, glyoxylate reductase | | Authors: | Duan, X, Hu, S, Zhou, P, Zhou, Y, Jiang, Z. | | Deposit date: | 2013-12-17 | | Release date: | 2014-12-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Characterization and crystal structure of a first fungal glyoxylate reductase from Paecilomyes thermophila
Enzyme.Microb.Technol., 60, 2014
|
|
6XQV
 
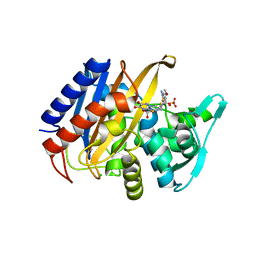 | | Crystal structure of the catalytic domain of PBP2 S310A from Neisseria gonorrhoeae in a pre-acylation complex with ceftriaxone | | Descriptor: | CHLORIDE ION, Ceftriaxone, Probable peptidoglycan D,D-transpeptidase PenA, ... | | Authors: | Fenton, B.A, Zhou, P, Davies, C. | | Deposit date: | 2020-07-10 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mutations in PBP2 from ceftriaxone-resistant Neisseria gonorrhoeae alter the dynamics of the beta 3-beta 4 loop to favor a low-affinity drug-binding state.
J.Biol.Chem., 297, 2021
|
|
6XQZ
 
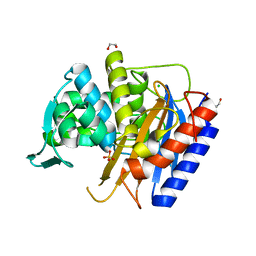 | | Crystal structure of the catalytic domain of PBP2 S310A from Neisseria gonorrhoeae at pH 7.5 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Peptidoglycan D,D-transpeptidase PenA, ... | | Authors: | Fenton, B.A, Zhou, P, Davies, C. | | Deposit date: | 2020-07-10 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Mutations in PBP2 from ceftriaxone-resistant Neisseria gonorrhoeae alter the dynamics of the beta 3-beta 4 loop to favor a low-affinity drug-binding state.
J.Biol.Chem., 297, 2021
|
|
6XQY
 
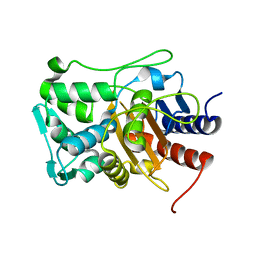 | |
6XQX
 
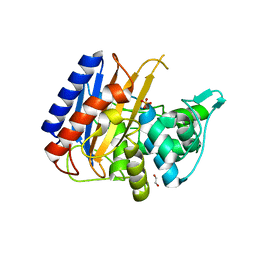 | | Crystal structure of the catalytic domain of PBP2 S310A from Neisseria gonorrhoeae with the H514A mutation at pH 7.5 | | Descriptor: | 1,2-ETHANEDIOL, Probable peptidoglycan D,D-transpeptidase PenA, SULFATE ION | | Authors: | Fenton, B.A, Zhou, P, Davies, C. | | Deposit date: | 2020-07-10 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mutations in PBP2 from ceftriaxone-resistant Neisseria gonorrhoeae alter the dynamics of the beta 3-beta 4 loop to favor a low-affinity drug-binding state.
J.Biol.Chem., 297, 2021
|
|
4IS9
 
 | | Crystal Structure of the Escherichia coli LpxC/L-161,240 complex | | Descriptor: | (4R)-2-(3,4-dimethoxy-5-propylphenyl)-N-hydroxy-4,5-dihydro-1,3-oxazole-4-carboxamide, ISOPROPYL ALCOHOL, SODIUM ION, ... | | Authors: | Lee, C.-J, Zhou, P. | | Deposit date: | 2013-01-16 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural Basis of the Promiscuous Inhibitor Susceptibility of Escherichia coli LpxC.
Acs Chem.Biol., 9, 2014
|
|
2MRM
 
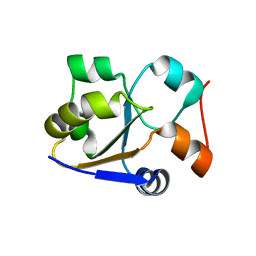 | |
7MK3
 
 | | Crystal structure of NPR1 | | Descriptor: | CHLORIDE ION, GLYCEROL, Regulatory protein NPR1, ... | | Authors: | Cheng, J, Wu, Q, Zhou, P. | | Deposit date: | 2021-04-21 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
2KIQ
 
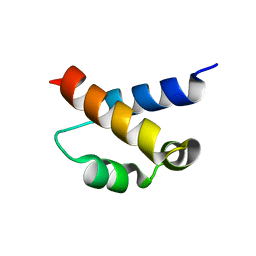 | | Solution structure of the FF Domain 2 of human transcription elongation factor CA150 | | Descriptor: | Transcription elongation regulator 1 | | Authors: | Zeng, J, Boyles, J, Tripathy, C, Yan, A, Zhou, P, Donald, B.R. | | Deposit date: | 2009-05-07 | | Release date: | 2009-07-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution protein structure determination starting with a global fold calculated from exact solutions to the RDC equations.
J.Biomol.Nmr, 45, 2009
|
|
2B5L
 
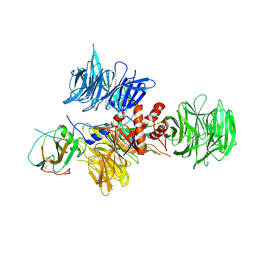 | | Crystal Structure of DDB1 In Complex with Simian Virus 5 V Protein | | Descriptor: | Nonstructural protein V, ZINC ION, damage-specific DNA binding protein 1 | | Authors: | Li, T, Chen, X, Garbutt, K.C, Zhou, P, Zheng, N. | | Deposit date: | 2005-09-28 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of DDB1 in complex with a paramyxovirus V protein: viral hijack of a propeller cluster in ubiquitin ligase.
Cell(Cambridge,Mass.), 124, 2006
|
|
2LSJ
 
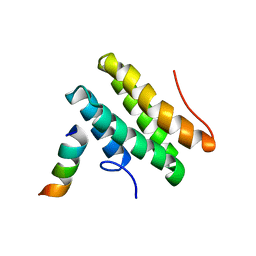 | |
2LSG
 
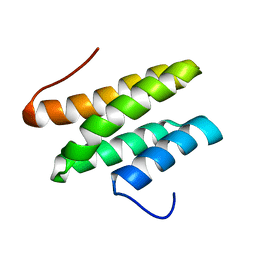 | |
6C59
 
 | | Chimeric Pol kappa RIR Rev1 C-terminal domain | | Descriptor: | ACETATE ION, Chimeric Pol kappa RIR Rev1 C-terminal domain | | Authors: | Wojtaszek, J.L, Zhou, P. | | Deposit date: | 2018-01-15 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A Small Molecule Targeting Mutagenic Translesion Synthesis Improves Chemotherapy.
Cell, 178, 2019
|
|
2JT2
 
 | | Solution Structure of the Aquifex aeolicus LpxC- CHIR-090 complex | | Descriptor: | N-{(1S,2R)-2-hydroxy-1-[(hydroxyamino)carbonyl]propyl}-4-{[4-(morpholin-4-ylmethyl)phenyl]ethynyl}benzamide, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Barb, A.W, Jiang, L, Raetz, C.R.H, Zhou, P. | | Deposit date: | 2007-07-18 | | Release date: | 2007-12-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the deacetylase LpxC bound to the antibiotic CHIR-090: Time-dependent inhibition and specificity in ligand binding
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
8E4A
 
 | | Pseudomonas LpxC in complex with LPC-233 | | Descriptor: | 4-(4-cyclopropylbuta-1,3-diyn-1-yl)-N-[(2S,3S)-4,4-difluoro-3-hydroxy-1-(hydroxyamino)-3-methyl-1-oxobutan-2-yl]benzamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Najeeb, J, Zhou, P. | | Deposit date: | 2022-08-17 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.034 Å) | | Cite: | Preclinical safety and efficacy characterization of an LpxC inhibitor against Gram-negative pathogens.
Sci Transl Med, 15, 2023
|
|
6C8C
 
 | | Chimeric Pol kappa RIR Rev1 C-terminal domain in complex with JHRE06 | | Descriptor: | 8-chloro-2-[(2,4-dichlorophenyl)amino]-3-(3-methylbutanoyl)-5-nitroquinolin-4(1H)-one, Chimeric protein of the Pol Kappa RIR helix and the Rev1 C-terminal domain | | Authors: | Najeeb, J, Zhou, P. | | Deposit date: | 2018-01-24 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Small Molecule Targeting Mutagenic Translesion Synthesis Improves Chemotherapy.
Cell, 178, 2019
|
|
6WII
 
 | | Crystal structure of the K. pneumoniae LpxH/JH-LPH-41 complex | | Descriptor: | 5-({[4-({4-[3-chloro-5-(trifluoromethyl)phenyl]piperazin-1-yl}sulfonyl)phenyl]carbamoyl}amino)-N-hydroxypentanamide, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Cochrane, C.S, Cho, J, Fenton, B.A, Zhou, P. | | Deposit date: | 2020-04-09 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Synthesis and evaluation of sulfonyl piperazine LpxH inhibitors.
Bioorg.Chem., 102, 2020
|
|
