6AL8
 
 | | Crystal structure HpiC1 Y101F/F138S | | 分子名称: | 1,2-ETHANEDIOL, 12-epi-hapalindole C/U synthase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | 著者 | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | 登録日 | 2017-08-07 | | 公開日 | 2018-03-07 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.641 Å) | | 主引用文献 | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
5YOX
 
 | | HD domain-containing protein YGK1(YGL101W) | | 分子名称: | HD domain-containing protein YGL101W, ZINC ION | | 著者 | Yang, J, Wang, F, Gao, Z, Zhou, K, Liu, Q. | | 登録日 | 2017-10-31 | | 公開日 | 2018-11-21 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | HD domain-containing protein YGK1(YGL101W)
To Be Published
|
|
6AL6
 
 | | Crystal structure HpiC1 in P42 space group | | 分子名称: | 12-epi-hapalindole C/U synthase, CALCIUM ION | | 著者 | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | 登録日 | 2017-08-07 | | 公開日 | 2018-03-07 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.088 Å) | | 主引用文献 | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
5Y37
 
 | | Crystal structure of GBS GAPDH | | 分子名称: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | 著者 | Jin, T, Zhou, K. | | 登録日 | 2017-07-28 | | 公開日 | 2018-04-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | High-resolution crystal structure of Streptococcus agalactiae glyceraldehyde-3-phosphate dehydrogenase.
Acta Crystallogr.,Sect.F, 74, 2018
|
|
5WXY
 
 | | Crystal structure of Microcystis aeruginosa PCC 7806 aspartate racemase in complex with L-aspartate | | 分子名称: | ASPARTIC ACID, McyF | | 著者 | Cao, D.D, Zhou, K, Jiang, Y.L, Zhou, C.Z. | | 登録日 | 2017-01-09 | | 公開日 | 2018-01-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.63 Å) | | 主引用文献 | Structural insights into the catalysis and substrate specificity of cyanobacterial aspartate racemase McyF.
Biochem.Biophys.Res.Commun., 514, 2019
|
|
5WXX
 
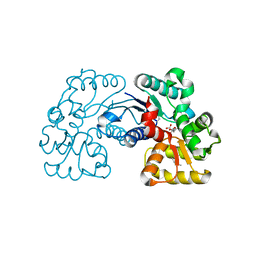 | | Crystal structure of Microcystis aeruginosa PCC 7806 aspartate racemase in complex with citrate | | 分子名称: | CITRIC ACID, McyF | | 著者 | Cao, D.D, Zhou, K, Jiang, Y.L, Zhou, C.Z. | | 登録日 | 2017-01-09 | | 公開日 | 2018-01-17 | | 最終更新日 | 2019-07-31 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structural insights into the catalysis and substrate specificity of cyanobacterial aspartate racemase McyF.
Biochem.Biophys.Res.Commun., 514, 2019
|
|
5WXZ
 
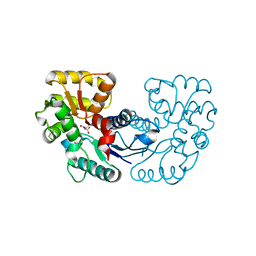 | | Crystal structure of Microcystis aeruginosa PCC 7806 aspartate racemase in complex with D-aspartate | | 分子名称: | D-ASPARTIC ACID, McyF | | 著者 | Cao, D.D, Zhou, K, Jiang, Y.L, Zhou, C.Z. | | 登録日 | 2017-01-09 | | 公開日 | 2018-01-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural insights into the catalysis and substrate specificity of cyanobacterial aspartate racemase McyF.
Biochem.Biophys.Res.Commun., 514, 2019
|
|
4K6N
 
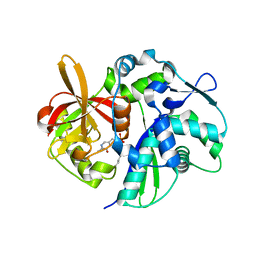 | | Crystal structure of yeast 4-amino-4-deoxychorismate lyase | | 分子名称: | Aminodeoxychorismate lyase, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Dai, Y.-N, Chi, C.-B, Zhou, K, Cheng, W, Jiang, Y.-L, Ren, Y.-M, Chen, Y, Zhou, C.-Z. | | 登録日 | 2013-04-16 | | 公開日 | 2013-07-10 | | 最終更新日 | 2013-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure and catalytic mechanism of yeast 4-amino-4-deoxychorismate lyase
J.Biol.Chem., 288, 2013
|
|
3J8Y
 
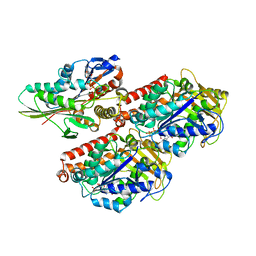 | | High-resolution structure of ATP analog-bound kinesin on microtubules | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Shang, Z, Zhou, K, Xu, C, Csencsits, R, Cochran, J.C, Sindelar, C.V. | | 登録日 | 2014-11-20 | | 公開日 | 2014-12-10 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (5 Å) | | 主引用文献 | High-resolution structures of kinesin on microtubules provide a basis for nucleotide-gated force-generation.
Elife, 3, 2014
|
|
6J3Q
 
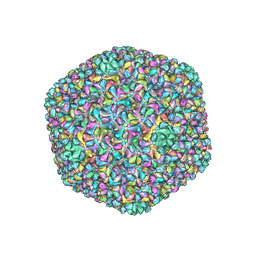 | | Capsid structure of a freshwater cyanophage Siphoviridae Mic1 | | 分子名称: | cement protein, major capsid protein | | 著者 | Jin, H, Jiang, Y.L, Yang, F, Zhang, J.T, Li, W.F, Zhou, K, Ju, J, Chen, Y, Zhou, C.Z. | | 登録日 | 2019-01-05 | | 公開日 | 2019-10-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.53 Å) | | 主引用文献 | Capsid Structure of a Freshwater Cyanophage Siphoviridae Mic1.
Structure, 27, 2019
|
|
6JOB
 
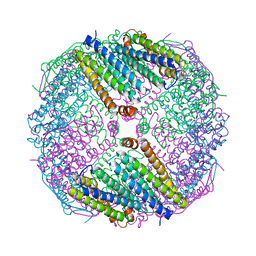 | | Ferritin variant with "GMG" motif | | 分子名称: | Ferritin heavy chain | | 著者 | Zheng, B.W, Zhou, K, Zhang, T, Lv, C, Wang, H, Zhao, G. | | 登録日 | 2019-03-20 | | 公開日 | 2020-03-25 | | 最終更新日 | 2024-03-27 | | 実験手法 | SOLUTION SCATTERING (2.93 Å), X-RAY DIFFRACTION | | 主引用文献 | Self-assembly of protein nanocage into designed 2D and 3D networks by grafting amyloidogenic motifs on the exterior surfaces.
To Be Published
|
|
7S3J
 
 | | Crystal Structure of AspB P450 in complex with brevianamide F substrates | | 分子名称: | (3S,8aS)-3-(1H-indol-3-ylmethyl)hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, AspB, GLYCEROL, ... | | 著者 | Newmister, S.A, Shende, V.V, Harris, N.R, Sanders, J.N, Khatri, Y, Movassaghi, M, Houk, K.N, Sherman, D.H. | | 登録日 | 2021-09-07 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Molecular Dynamics Simulations Guide Chimeragenesis and Engineered Control of Chemoselectivity in Diketopiperazine Dimerases.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
3J0A
 
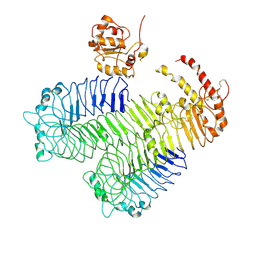 | | Homology model of human Toll-like receptor 5 fitted into an electron microscopy single particle reconstruction | | 分子名称: | Toll-like receptor 5, alpha-L-fucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Modis, Y, Zhou, K, Kanai, R, Lee, P, Wang, H.W. | | 登録日 | 2011-06-02 | | 公開日 | 2011-12-28 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (26 Å) | | 主引用文献 | Toll-like receptor 5 forms asymmetric dimers in the absence of flagellin.
J.Struct.Biol., 177, 2012
|
|
4R0M
 
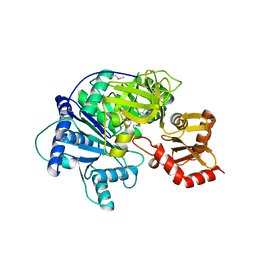 | | Structure of McyG A-PCP complexed with phenylalanyl-adenylate | | 分子名称: | ADENOSINE-5'-[PHENYLALANINYL-PHOSPHATE], McyG protein | | 著者 | Tan, X.F, Dai, Y.N, Zhou, K, Jiang, Y.L, Ren, Y.M, Chen, Y.X, Zhou, C.Z. | | 登録日 | 2014-08-01 | | 公開日 | 2015-04-15 | | 最終更新日 | 2019-12-18 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Structure of the adenylation-peptidyl carrier protein didomain of the Microcystis aeruginosa microcystin synthetase McyG.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3J8X
 
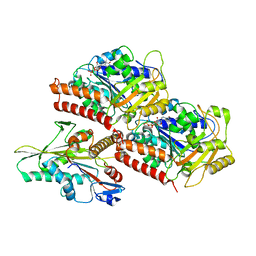 | | High-resolution structure of no-nucleotide kinesin on microtubules | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-1 heavy chain, ... | | 著者 | Shang, Z, Zhou, K, Xu, C, Csencsits, R, Cochran, J.C, Sindelar, C.V. | | 登録日 | 2014-11-20 | | 公開日 | 2014-12-10 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (5 Å) | | 主引用文献 | High-resolution structures of kinesin on microtubules provide a basis for nucleotide-gated force-generation.
Elife, 3, 2014
|
|
4QSG
 
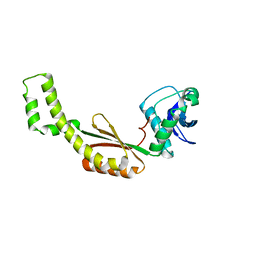 | | Crystal structure of gas vesicle protein GvpF from Microcystis aeruginosa | | 分子名称: | Gas vesicle protein | | 著者 | Xu, B.Y, Dai, Y.N, Zhou, K, Ren, Y.M, Liu, Y.T, Chen, Y, Zhou, C.Z. | | 登録日 | 2014-07-03 | | 公開日 | 2014-11-12 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure of the gas vesicle protein GvpF from the cyanobacterium Microcystis aeruginosa.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6J4A
 
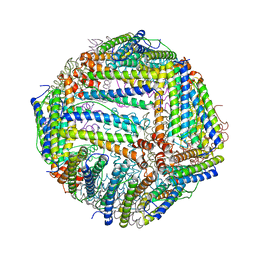 | |
3UZ5
 
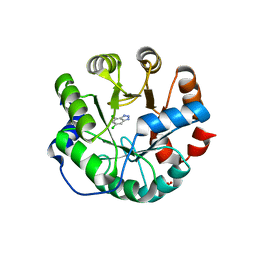 | | Designed protein KE59 R13 3/11H | | 分子名称: | 5,7-dichloro-1H-benzotriazole, Kemp eliminase KE59 R13 3/11H, PHOSPHATE ION, ... | | 著者 | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2011-12-07 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UXD
 
 | | Designed protein KE59 R1 7/10H with dichlorobenzotriazole (DBT) | | 分子名称: | 5,7-dichloro-1H-benzotriazole, Kemp eliminase KE59 R1 7/10H, PHOSPHATE ION | | 著者 | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2011-12-05 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UY7
 
 | | Designed protein KE59 R1 7/10H with G130S mutation | | 分子名称: | Kemp eliminase KE59 R1 7/10H, SODIUM ION, SULFATE ION | | 著者 | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2011-12-06 | | 公開日 | 2012-06-27 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6J4M
 
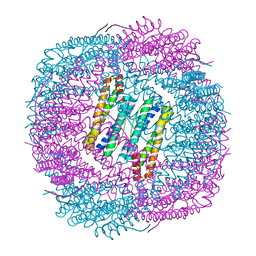 | | Thermal treated soybean seed H-2 ferritin | | 分子名称: | Ferritin, MAGNESIUM ION | | 著者 | Zhang, X, Zang, J, Chen, H, Zhou, K, Zhao, G. | | 登録日 | 2019-01-09 | | 公開日 | 2019-09-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.598 Å) | | 主引用文献 | Thermostability of protein nanocages: the effect of natural extra peptide on the exterior surface.
Rsc Adv, 9, 2019
|
|
5WPP
 
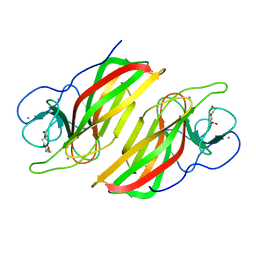 | | Crystal structure HpiC1 W73M/K132M | | 分子名称: | 12-epi-hapalindole C/U synthase, CALCIUM ION, TETRAETHYLENE GLYCOL, ... | | 著者 | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | 登録日 | 2017-08-07 | | 公開日 | 2018-03-07 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
3UXA
 
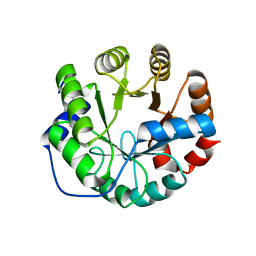 | | Designed protein KE59 R1 7/10H | | 分子名称: | Kemp eliminase KE59 R1 7/10H, PHOSPHATE ION | | 著者 | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2011-12-05 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UZJ
 
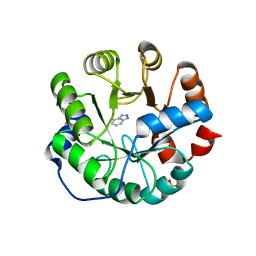 | | Designed protein KE59 R13 3/11H with benzotriazole | | 分子名称: | 1H-benzotriazole, Kemp eliminase KE59 R13 3/11H, PHOSPHATE ION | | 著者 | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2011-12-07 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UY8
 
 | | Designed protein KE59 R5_11/5F | | 分子名称: | Kemp eliminase KE59 R5_11/5F, SULFATE ION | | 著者 | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2011-12-06 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
