4QLX
 
 | | Crystal structure of CLA-ER with product binding | | Descriptor: | 10-oxooctadecanoic acid, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Hou, F, Miyakawa, T, Tanokura, M. | | Deposit date: | 2014-06-13 | | Release date: | 2015-02-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and reaction mechanism of a novel enone reductase.
Febs J., 282, 2015
|
|
4QLY
 
 | | Crystal structure of CLA-ER, a novel enone reductase catalyzing a key step of a gut-bacterial fatty acid saturation metabolism, biohydrogenation | | Descriptor: | Enone reductase CLA-ER, FLAVIN MONONUCLEOTIDE | | Authors: | Hou, F, Miyakawa, T, Tanokura, M. | | Deposit date: | 2014-06-13 | | Release date: | 2015-02-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Structure and reaction mechanism of a novel enone reductase.
Febs J., 282, 2015
|
|
3WDS
 
 | |
7CZ5
 
 | | Cryo-EM structure of the human growth hormone-releasing hormone receptor-Gs protein complex | | Descriptor: | CHOLESTEROL, Growth hormone-releasing hormone receptor,growth hormone-releasing hormone receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhou, F, Zhang, H, Cong, Z, Zhao, L, Zhou, Q, Mao, C, Cheng, X, Shen, D, Cai, X, Ma, C, Wang, Y, Dai, A, Zhou, Y, Sun, W, Zhao, F, Zhao, S, Jiang, H, Jiang, Y, Yang, D, Xu, H.E, Zhang, Y, Wang, M. | | Deposit date: | 2020-09-07 | | Release date: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis for activation of the growth hormone-releasing hormone receptor.
Nat Commun, 11, 2020
|
|
6EBA
 
 | | Crystal Structure of A Bacterial Homolog to Human Lysosomal Transporter, Spinster, in Inward-facing And Unoccupied Conformation | | Descriptor: | Major facilitator family transporter | | Authors: | Zhou, F, Yao, D, Rao, B, Zhang, L, Cao, Y. | | Deposit date: | 2018-08-06 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.812 Å) | | Cite: | Crystal structure of a bacterial homolog to human lysosomal transporter, spinster
Sci Bull (Beijing), 64, 2019
|
|
7DW9
 
 | |
8OS6
 
 | | Structure of a GFRA1/GDNF LICAM complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Houghton, F.M, Adams, S.E, Briggs, D.C, McDonald, N.Q. | | Deposit date: | 2023-04-18 | | Release date: | 2023-11-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Architecture and regulation of a GDNF-GFR alpha 1 synaptic adhesion assembly.
Nat Commun, 14, 2023
|
|
6E8J
 
 | | Crystal Structure of A Bacterial Homolog to Human Lysosomal Transporter, Spinster, in Inward-facing And Occupied Conformation | | Descriptor: | Major facilitator family transporter | | Authors: | Zhou, F, Yao, D, Rao, B, Zhang, L, Cao, Y. | | Deposit date: | 2018-07-30 | | Release date: | 2019-08-14 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (3.091 Å) | | Cite: | Crystal structure of a bacterial homolog to human lysosomal transporter, spinster
Sci Bull (Beijing), 64, 2019
|
|
6E9C
 
 | | Selenomethionine Derivative Structure of A Bacterial Homolog to Human Lysosomal Transporter, Spinster | | Descriptor: | Major facilitator family transporter | | Authors: | Zhou, F, Yao, D, Rao, B, Zhang, L, Cao, Y. | | Deposit date: | 2018-07-31 | | Release date: | 2019-08-14 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a bacterial homolog to human lysosomal transporter, spinster
Sci Bull (Beijing), 64, 2019
|
|
5Z33
 
 | |
5OJR
 
 | | YCF48 bound to D1 peptide | | Descriptor: | Photosystem II protein D1 3, Ycf48-like protein | | Authors: | Michoux, F, Nixon, P.J, Murray, J.W, Bialek, W, Thieulin-Pardo, G. | | Deposit date: | 2017-07-23 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Ycf48 involved in the biogenesis of the oxygen-evolving photosystem II complex is a seven-bladed beta-propeller protein.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5OJ3
 
 | | YCF48 from Cyanidioschyzon merolae | | Descriptor: | Photosystem II stability/assembly factor HCF136 | | Authors: | Michoux, F, Murray, J.W, Nixon, P.J. | | Deposit date: | 2017-07-20 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.982 Å) | | Cite: | Ycf48 involved in the biogenesis of the oxygen-evolving photosystem II complex is a seven-bladed beta-propeller protein.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5OJ5
 
 | | YCF48 bound to D1 peptide | | Descriptor: | PHE-PRO-LEU-ASP-LEU-ALA, Ycf48-like protein | | Authors: | Michoux, F, Nixon, P.J, Murray, J.W. | | Deposit date: | 2017-07-20 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Ycf48 involved in the biogenesis of the oxygen-evolving photosystem II complex is a seven-bladed beta-propeller protein.
Proc.Natl.Acad.Sci.USA, 115, 2018
|
|
5OJP
 
 | | YCF48 bound to D1 peptide | | Descriptor: | Ycf48-like protein | | Authors: | Michoux, F, Nixon, P.J, Murray, J.W. | | Deposit date: | 2017-07-22 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Ycf48 involved in the biogenesis of the oxygen-evolving photosystem II complex is a seven-bladed beta-propeller protein.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2XB3
 
 | | The Structure of Cyanobacterial PsbP | | Descriptor: | PSBP PROTEIN, ZINC ION | | Authors: | Michoux, F, Takasaka, K, Nixon, P, Murray, J.W. | | Deposit date: | 2010-04-03 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Structure of Cyanop at 2.8A: Implications for the Evolution and Function of the Psbp Subunit of Photosystem II.
Biochemistry, 49, 2010
|
|
2XBG
 
 | |
2Y6X
 
 | | Structure of Psb27 from Thermosynechococcus elongatus | | Descriptor: | CHLORIDE ION, PHOTOSYSTEM II 11 KD PROTEIN | | Authors: | Michoux, F, Takasaka, K, Boehm, M, Nixon, P.J, Murray, J.W. | | Deposit date: | 2011-01-27 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Psb27 Assembly Factor at 1.6A: Implications for Binding to Photosystem II.
Photosynth.Res., 110, 2012
|
|
3ZSU
 
 | | Structure of the CyanoQ protein from Thermosynechococcus elongatus | | Descriptor: | SULFATE ION, TLL2057 PROTEIN | | Authors: | Michoux, F, Takasaka, K, Nixon, P.J, Murray, J.W. | | Deposit date: | 2011-06-30 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Cyanoq from the Thermophilic Cyanobacterium Thermosynechococcus Elongatus and Detection in Isolated Photosystem II Complexes.
Photosynth.Res., 122, 2014
|
|
7L97
 
 | | Crystal structure of STAMBPL1 in complex with an engineered binder | | Descriptor: | 1,2-ETHANEDIOL, AMSH-like protease, SULFATE ION, ... | | Authors: | Guo, Y, Dong, A, Hou, F, Li, Y, Zhang, W, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-01-02 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and functional characterization of ubiquitin variant inhibitors for the JAMM-family deubiquitinases STAMBP and STAMBPL1.
J.Biol.Chem., 297, 2021
|
|
5YPX
 
 | | Crystal structure of calaxin with magnesium | | Descriptor: | Calaxin, MAGNESIUM ION | | Authors: | Shojima, T, Hou, F, Takahashi, Y, Okai, M, Mizuno, K, Inaba, K, Miyakawa, T, Tanokura, M. | | Deposit date: | 2017-11-04 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structure of a Ca2+-dependent regulator of flagellar motility reveals the open-closed structural transition
Sci Rep, 8, 2018
|
|
5X9A
 
 | | Crystal structure of calaxin with calcium | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Calaxin | | Authors: | Shojima, T, Hou, F, Takahashi, Y, Okai, M, Mizuno, K, Inaba, K, Miyakawa, T, Tanokura, M. | | Deposit date: | 2017-03-06 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of a Ca2+-dependent regulator of flagellar motility reveals the open-closed structural transition
Sci Rep, 8, 2018
|
|
3WGC
 
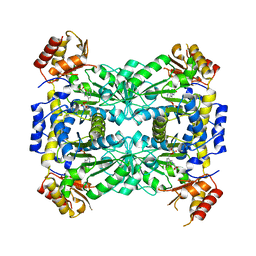 | | Aeromonas jandaei L-allo-threonine aldolase H128Y/S292R double mutant | | Descriptor: | L-allo-threonine aldolase, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE] | | Authors: | Qin, H.M, Imai, F.L, Miyakawa, T, Kataoka, M, Okai, M, Ohtsuka, J, Hou, F, Nagata, K, Shimizu, S, Tanokura, M. | | Deposit date: | 2013-08-03 | | Release date: | 2014-07-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | L-allo-Threonine aldolase with an H128Y/S292R mutation from Aeromonas jandaei DK-39 reveals the structural basis of changes in substrate stereoselectivity.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WLX
 
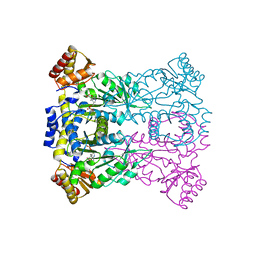 | | Crystal structure of low-specificity L-threonine aldolase from Escherichia coli | | Descriptor: | Low specificity L-threonine aldolase, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE] | | Authors: | Qin, H.-M, Imai, F.L, Miyakawa, T, Kataoka, M, Okai, M, Hou, F, Ohtsuka, J, Nagata, K, Shimizu, S, Tanokura, M. | | Deposit date: | 2013-11-15 | | Release date: | 2014-12-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure analysis of L-threonine aldolase from Escherichia coli unravels the low-specificity and thermostability
To be Published
|
|
3WGB
 
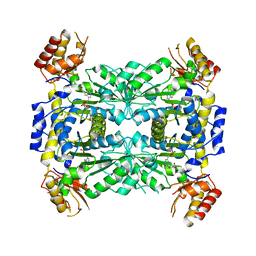 | | Crystal structure of aeromonas jandaei L-allo-threonine aldolase | | Descriptor: | GLYCINE, L-allo-threonine aldolase, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE] | | Authors: | Qin, H.M, Imai, F.L, Miyakawa, T, Kataoka, M, Okai, M, Ohtsuka, J, Hou, F, Nagata, K, Shimizu, S, Tanokura, M. | | Deposit date: | 2013-08-03 | | Release date: | 2014-07-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | L-allo-Threonine aldolase with an H128Y/S292R mutation from Aeromonas jandaei DK-39 reveals the structural basis of changes in substrate stereoselectivity.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WWJ
 
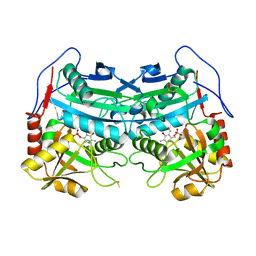 | | Crystal structure of an engineered sitagliptin-producing transaminase, ATA-117-Rd11 | | Descriptor: | (R)-amine transaminase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Guan, L.J, Ohtsuka, J, Okai, M, Miyakawa, T, Mase, T, Zhi, Y, Hou, F, Ito, N, Yasohara, Y, Tanokura, M. | | Deposit date: | 2014-06-18 | | Release date: | 2015-08-12 | | Last modified: | 2018-11-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A new target region for changing the substrate specificity of amine transaminases.
Sci Rep, 5, 2015
|
|
