6JD0
 
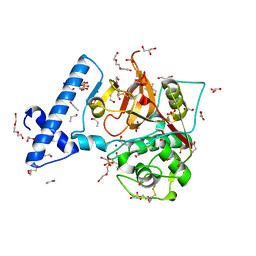 | | Structure of mutant human cathepsin L, engineered for GAG binding | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cathepsin L1, ... | | Authors: | Choudhury, D, Biswas, S. | | Deposit date: | 2019-01-30 | | Release date: | 2020-02-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Structure-guided protein engineering of human cathepsin L for efficient collagenolytic activity.
Protein Eng.Des.Sel., 34, 2021
|
|
1U0I
 
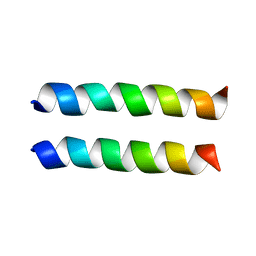 | | IAAL-E3/K3 heterodimer | | Descriptor: | IAAL-E3, IAAL-K3 | | Authors: | Lindhout, D.A, Litowski, J.R, Mercier, P, Hodges, R.S, Sykes, B.D. | | Deposit date: | 2004-07-13 | | Release date: | 2004-10-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of a highly stable de novo heterodimeric coiled-coil
Biopolymers, 75, 2004
|
|
6N2E
 
 | |
1W1Y
 
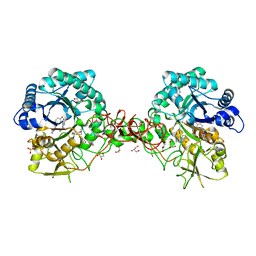 | | Crystal structure of S. marcescens chitinase B in complex with the cyclic dipeptide inhibitor cyclo-(L-Tyr-L-Pro) at 1.85 A resolution | | Descriptor: | CHITINASE B, CYCLO-(L-TYROSINE-L-PROLINE) INHIBITOR, GLYCEROL, ... | | Authors: | Houston, D.R, Synstad, B, Eijsink, V.G.H, Eggleston, I, Van Aalten, D.M.F. | | Deposit date: | 2004-06-24 | | Release date: | 2005-01-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Based Exploration of Cyclic Dipeptide Chitinase Inhibitors
J.Med.Chem., 47, 2004
|
|
1W1T
 
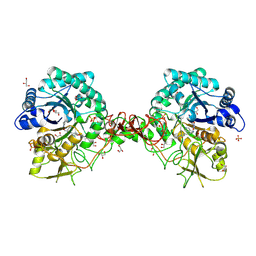 | | Crystal structure of S. marcescens chitinase B in complex with the cyclic dipeptide inhibitor cyclo-(His-L-Pro) at 1.9 A resolution | | Descriptor: | CHITINASE B, CYCLO-(L-HISTIDINE-L-PROLINE) INHIBITOR, GLYCEROL, ... | | Authors: | Houston, D.R, Synstad, B, Eijsink, V.G.H, Eggleston, I, van Aalten, D.M.F. | | Deposit date: | 2004-06-24 | | Release date: | 2005-01-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Exploration of Cyclic Dipeptide Chitinase Inhibitors
J.Med.Chem., 47, 2004
|
|
8PTI
 
 | | CRYSTAL STRUCTURE OF A Y35G MUTANT OF BOVINE PANCREATIC TRYPSIN INHIBITOR | | Descriptor: | BOVINE PANCREATIC TRYPSIN INHIBITOR | | Authors: | Housset, D, Kim, K.-S, Fuchs, J, Woodward, C, Wlodawer, A. | | Deposit date: | 1990-12-17 | | Release date: | 1991-04-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a Y35G mutant of bovine pancreatic trypsin inhibitor.
J.Mol.Biol., 220, 1991
|
|
1W1V
 
 | | Crystal structure of S. marcescens chitinase B in complex with the cyclic dipeptide inhibitor cyclo-(L-Arg-L-Pro) at 1.85 A resolution | | Descriptor: | CHITINASE B, CYCLO-(L-ARGININE-L-PROLINE) INHIBITOR, GLYCEROL, ... | | Authors: | Houston, D.R, Synstad, B, Eijsink, V.G.H, Eggleston, I, Van Aalten, D.M.F. | | Deposit date: | 2004-06-24 | | Release date: | 2005-01-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Based Exploration of Cyclic Dipeptide Chitinase Inhibitors
J.Med.Chem., 47, 2004
|
|
1W1P
 
 | | Crystal structure of S. marcescens chitinase B in complex with the cyclic dipeptide inhibitor cyclo-(Gly-L-Pro) at 2.1 A resolution | | Descriptor: | CHITINASE B, CYCLO-(GLYCINE-L-PROLINE) INHIBITOR, GLYCEROL, ... | | Authors: | Houston, D.R, Synstad, B, Eijsink, V.G.H, Eggleston, I, Van Aalten, D.M.F. | | Deposit date: | 2004-06-23 | | Release date: | 2005-01-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Exploration of Cyclic Dipeptide Chitinase Inhibitors
J.Med.Chem., 47, 2004
|
|
1OZS
 
 | |
1KB5
 
 | | MURINE T-CELL RECEPTOR VARIABLE DOMAIN/FAB COMPLEX | | Descriptor: | ANTIBODY DESIRE-1, KB5-C20 T-CELL ANTIGEN RECEPTOR | | Authors: | Housset, D, Mazza, G, Gregoire, C, Piras, C, Malissen, B, Fontecilla-Camps, J.C. | | Deposit date: | 1997-04-06 | | Release date: | 1998-04-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The three-dimensional structure of a T-cell antigen receptor V alpha V beta heterodimer reveals a novel arrangement of the V beta domain.
EMBO J., 16, 1997
|
|
1FSC
 
 | |
1FDS
 
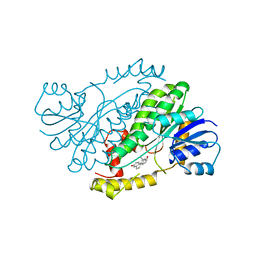 | | HUMAN 17-BETA-HYDROXYSTEROID-DEHYDROGENASE TYPE 1 COMPLEXED WITH 17-BETA-ESTRADIOL | | Descriptor: | 17-BETA-HYDROXYSTEROID-DEHYDROGENASE, ESTRADIOL | | Authors: | Housset, D, Breton, R, Mazza, C, Fontecilla-Camps, J.-C. | | Deposit date: | 1996-06-28 | | Release date: | 1997-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of a complex of human 17beta-hydroxysteroid dehydrogenase with estradiol and NADP+ identifies two principal targets for the design of inhibitors.
Structure, 4, 1996
|
|
1O6I
 
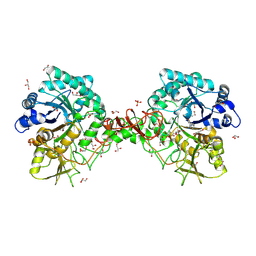 | | Chitinase B from Serratia marcescens complexed with the catalytic intermediate mimic cyclic dipeptide CI4. | | Descriptor: | Chitinase, GLYCEROL, SULFATE ION, ... | | Authors: | Houston, D.R, Eggleston, I, Synstad, B, Eijsink, V.G.H, van Aalten, D.M.F. | | Deposit date: | 2002-10-03 | | Release date: | 2003-03-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The cyclic dipeptide CI-4 [cyclo-(l-Arg-d-Pro)] inhibits family 18 chitinases by structural mimicry of a reaction intermediate.
Biochem. J., 368, 2002
|
|
1FDT
 
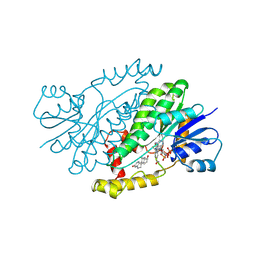 | | HUMAN 17-BETA-HYDROXYSTEROID-DEHYDROGENASE TYPE 1 COMPLEXED WITH ESTRADIOL AND NADP+ | | Descriptor: | 17-BETA-HYDROXYSTEROID-DEHYDROGENASE, ESTRADIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Housset, D, Breton, R, Mazza, C, Fontecilla-Camps, J.-C. | | Deposit date: | 1996-06-28 | | Release date: | 1997-02-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of a complex of human 17beta-hydroxysteroid dehydrogenase with estradiol and NADP+ identifies two principal targets for the design of inhibitors.
Structure, 4, 1996
|
|
3B9C
 
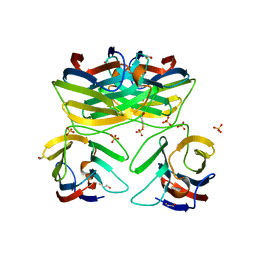 | | Crystal Structure of Human GRP CRD | | Descriptor: | BETA-MERCAPTOETHANOL, HSPC159, SULFATE ION | | Authors: | Zhou, D, Ge, H.H, Niu, L.W, Teng, M.K. | | Deposit date: | 2007-11-05 | | Release date: | 2008-03-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the C-terminal conserved domain of human GRP, a galectin-related protein, reveals a function mode different from those of galectins.
Proteins, 71, 2008
|
|
1HJW
 
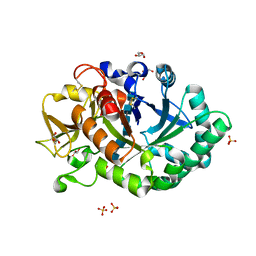 | | Crystal structure of hcgp-39 in complex with chitin octamer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Houston, D.R, Recklies, A.D, Krupa, J.C, Van Aalten, D.M.F. | | Deposit date: | 2003-02-28 | | Release date: | 2003-03-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Ligand-Induced Conformational Change of the 39-kDa Glycoprotein from Human Articular Chondrocytes
J.Biol.Chem., 278, 2003
|
|
4XAE
 
 | | Structure of Feruloyl-CoA 6-hydroxylase (F6H) from Arabidopsis thaliana | | Descriptor: | Feruloyl CoA ortho-hydroxylase 1, SODIUM ION | | Authors: | Zhou, D, Kandavelu, P, Zhang, H, Wang, B.C, Rose, J, Yan, Y. | | Deposit date: | 2014-12-14 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.769 Å) | | Cite: | Structural Insights into Substrate Specificity of Feruloyl-CoA 6'-Hydroxylase from Arabidopsis thaliana.
Sci Rep, 5, 2015
|
|
1H0I
 
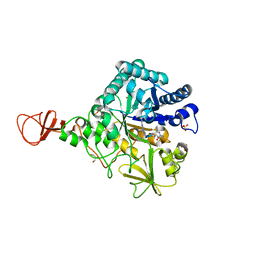 | | Complex of a chitinase with the natural product cyclopentapeptide argifin from Gliocladium | | Descriptor: | ARGIFIN, CHITINASE B, GLYCEROL, ... | | Authors: | Houston, D.R, Shiomi, K, Arai, N, Omura, S, Peter, M.G, Turberg, A, Synstad, B, Eijsink, V.G.H, Aalten, D.M.F. | | Deposit date: | 2002-06-19 | | Release date: | 2002-06-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High Resolution Inhibited Complexes of a Chitinase with Natural Product Cyclopentapeptides - Peptide Mimicry of a Carbohydrate Substrate
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
6FCM
 
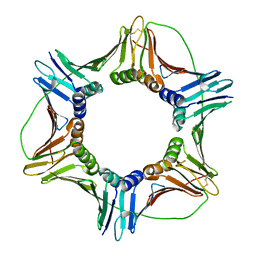 | | Crystal structure of human PCNA | | Descriptor: | Proliferating cell nuclear antigen | | Authors: | Housset, D, Frachet, P. | | Deposit date: | 2017-12-20 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Cytosolic PCNA interacts with p47phox and controls NADPH oxidase NOX2 activation in neutrophils.
J.Exp.Med., 216, 2019
|
|
6FCN
 
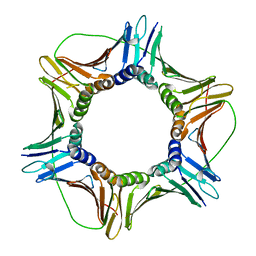 | |
6TA2
 
 | | Human NAMPT in complex with nicotinic acid mononucleotide and phosphate | | Descriptor: | CHLORIDE ION, GLYCEROL, NICOTINATE MONONUCLEOTIDE, ... | | Authors: | Houry, D, Raasakka, A, Kursula, P, Ziegler, M. | | Deposit date: | 2019-10-29 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Identification of structural determinants of NAMPT activity and substrate selectivity
To Be Published
|
|
6TAC
 
 | | Human NAMPT deletion mutant in complex with nicotinamide mononucleotide, pyrophosphate, and Mg2+ | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Houry, D, Raasakka, A, Kursula, P, Ziegler, M. | | Deposit date: | 2019-10-29 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of structural determinants of NAMPT activity and substrate selectivity
To Be Published
|
|
7PS3
 
 | | Crystal structure of antibody Beta-32 Fab | | Descriptor: | Beta-32 heavy chain, Beta-32 light chain, CHLORIDE ION, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-09-22 | | Release date: | 2021-12-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7PS0
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 beta variant spike glycoprotein in complex with beta-24 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-24 heavy chain, Beta-24 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-09-22 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7PS7
 
 | |
