4WHM
 
 | | Crystal structure of UDP-glucose: anthocyanidin 3-O-glucosyltransferase in complex with UDP | | Descriptor: | ACETATE ION, GLYCEROL, UDP-glucose:anthocyanidin 3-O-glucosyltransferase, ... | | Authors: | Hiromoto, T, Honjo, E, Tamada, T, Kuroki, R. | | Deposit date: | 2014-09-23 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Structural basis for acceptor-substrate recognition of UDP-glucose: anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea
Protein Sci., 24, 2015
|
|
3WC4
 
 | | Crystal structure of UDP-glucose: anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea | | Descriptor: | ACETATE ION, GLYCEROL, UDP-glucose:anthocyanidin 3-O-glucosyltransferase | | Authors: | Hiromoto, T, Honjo, E, Tamada, T, Kuroki, R. | | Deposit date: | 2013-05-24 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of UDP-glucose:anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea
J.SYNCHROTRON RADIAT., 20, 2013
|
|
4REN
 
 | | Crystal structure of UDP-glucose: anthocyanidin 3-O-glucosyltransferase in complex with petunidin | | Descriptor: | 2-(3,4-dihydroxy-5-methoxyphenyl)-3,5,7-trihydroxychromenium, GLYCEROL, UDP-glucose:anthocyanidin 3-O-glucosyltransferase | | Authors: | Hiromoto, T, Honjo, E, Tamada, T, Kuroki, R. | | Deposit date: | 2014-09-23 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Structural basis for acceptor-substrate recognition of UDP-glucose: anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea
Protein Sci., 24, 2015
|
|
4REM
 
 | | Crystal structure of UDP-glucose: anthocyanidin 3-O-glucosyltransferase in complex with delphinidin | | Descriptor: | 3,5,7-trihydroxy-2-(3,4,5-trihydroxyphenyl)chromenium, GLYCEROL, UDP-glucose:anthocyanidin 3-O-glucosyltransferase | | Authors: | Hiromoto, T, Honjo, E, Tamada, T, Kuroki, R. | | Deposit date: | 2014-09-23 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis for acceptor-substrate recognition of UDP-glucose: anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea
Protein Sci., 24, 2015
|
|
4REL
 
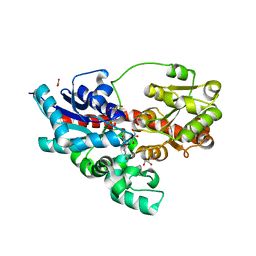 | | Crystal structure of UDP-glucose: anthocyanidin 3-O-glucosyltransferase in complex with kaempferol | | Descriptor: | 3,5,7-TRIHYDROXY-2-(4-HYDROXYPHENYL)-4H-CHROMEN-4-ONE, ACETATE ION, GLYCEROL, ... | | Authors: | Hiromoto, T, Honjo, E, Tamada, T, Kuroki, R. | | Deposit date: | 2014-09-23 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.754 Å) | | Cite: | Structural basis for acceptor-substrate recognition of UDP-glucose: anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea
Protein Sci., 24, 2015
|
|
1J1R
 
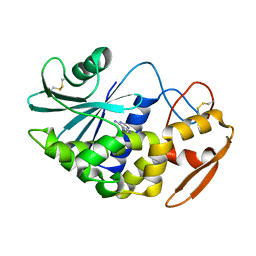 | | Structure of Pokeweed Antiviral Protein from Seeds (PAP-S1) Complexed with Adenine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENINE, Antiviral Protein S | | Authors: | Watanabe, K, Sato, E, Honjo, E, Motoshima, H, Kurokawa, H, Mikami, B, Monzingo, A.F, Robertus, J.D, Fujii, H, Hidaka, A. | | Deposit date: | 2002-12-14 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Pokweed Antiviral Protein from Seeds (PAP-S1) at 1.8 Angstrom Resolution
To be published
|
|
1J1Q
 
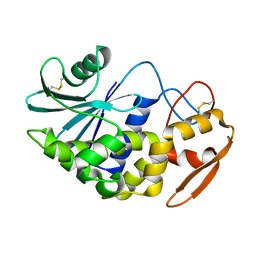 | | Structure of Pokeweed Antiviral Protein from Seeds (PAP-S1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antiviral protein S | | Authors: | Watanabe, K, Sato, E, Honjo, E, Motoshima, H, Kurokawa, H, Mikami, B, Monzingo, A.F, Robertus, J.D, Fujii, H, Hidaka, A. | | Deposit date: | 2002-12-14 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Pokweed Antiviral Protein from Seeds (PAP-S1) at 1.8 Angstrom Resolution
To be published
|
|
1J1S
 
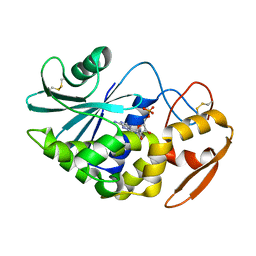 | | Pokeweed Antiviral Protein from Seeds (PAP-S1) Complexed with Formycin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antiviral Protein S, FORMYCIN-5'-MONOPHOSPHATE | | Authors: | Watanabe, K, Sato, E, Honjo, E, Motoshima, H, Kurokawa, H, Mikami, B, Monzingo, A.F, Robertus, J.D, Fujii, H, Hidaka, A. | | Deposit date: | 2002-12-14 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Pokweed Antiviral Protein from Seeds (PAP-S1) at 1.8 Angstrom Resolution
To be published
|
|
3CQA
 
 | | Crystal structure of human fibroblast growth factor-1 with mutations Glu81Ala and Lys101Ala | | Descriptor: | FORMIC ACID, Heparin-binding growth factor 1, SULFATE ION | | Authors: | Meher, A.K, Honjo, E, Kuroki, R, Lee, J, Somasundaram, T, Blaber, M. | | Deposit date: | 2008-04-02 | | Release date: | 2009-04-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering an improved crystal contact across a solvent-mediated interface of human fibroblast growth factor 1.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
2ZKH
 
 | | Human thrombopoietin neutralizing antibody TN1 FAB | | Descriptor: | Monoclonal TN1 FAB heavy chain, Monoclonal TN1 FAB light chain | | Authors: | Arai, S, Tamada, T, Honjo, E, Maeda, Y, Kuroki, R. | | Deposit date: | 2008-03-21 | | Release date: | 2009-03-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | An insight into the thermodynamic characteristics of human thrombopoietin complexation with TN1 antibody.
Protein Sci., 25, 2016
|
|
3CRG
 
 | | Crystal structure of human fibroblast growth factor-1 with mutations Glu81Ala, Glu82Asn and Lys101Ala | | Descriptor: | FORMIC ACID, GLYCEROL, Heparin-binding growth factor 1, ... | | Authors: | Meher, A.K, Honjo, E, Kuroki, R, Lee, J, Somasundaram, T, Blaber, M. | | Deposit date: | 2008-04-07 | | Release date: | 2009-02-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Engineering an improved crystal contact across a solvent-mediated interface of human fibroblast growth factor 1.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3CRI
 
 | | Crystal structure of human fibroblast growth factor-1 with mutations Glu81Ser, Glu82Asn and Lys101Ala | | Descriptor: | FORMIC ACID, Heparin-binding growth factor 1, SULFATE ION | | Authors: | Meher, A.K, Honjo, E, Kuroki, R, Lee, J, Somasundaram, T, Blaber, M. | | Deposit date: | 2008-04-07 | | Release date: | 2009-02-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Engineering an improved crystal contact across a solvent-mediated interface of human fibroblast growth factor 1.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3CRH
 
 | | Crystal structure of human fibroblast growth factor-1 with mutations Glu81Ser and Lys101Ala | | Descriptor: | Heparin-binding growth factor 1, SULFATE ION | | Authors: | Meher, A.K, Honjo, E, Kuroki, R, Lee, J, Somasundaram, T, Blaber, M. | | Deposit date: | 2008-04-07 | | Release date: | 2009-02-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Engineering an improved crystal contact across a solvent-mediated interface of human fibroblast growth factor 1.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
