2ANJ
 
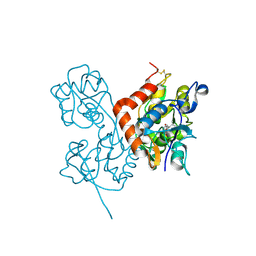 | | Crystal Structure of the Glur2 Ligand Binding Core (S1S2J-Y450W) Mutant in Complex With the Partial Agonist Kainic Acid at 2.1 A Resolution | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 2 | | Authors: | Holm, M.M, Naur, P, Vestergaard, B, Geballe, M.T, Gajhede, M, Kastrup, J.S, Traynelis, S.F, Egebjerg, J. | | Deposit date: | 2005-08-11 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Binding Site Tyrosine Shapes Desensitization Kinetics and Agonist Potency at GluR2: a mutagenic, kinetic, and crystallographic study
J.Biol.Chem., 280, 2005
|
|
8G6J
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (GA state 2) | | Descriptor: | (3R,6R,9S,12S,15S,18S,20R,24aR)-6-[(2S)-butan-2-yl]-3,12-bis[(1R)-1-hydroxy-2-methylpropyl]-8,9,11,17,18-pentamethyl-15-[(2S)-2-methylbutyl]hexadecahydropyrido[1,2-a][1,4,7,10,13,16,19]heptaazacyclohenicosine-1,4,7,10,13,16,19(21H)-heptone, (3beta)-O~3~-[(2R)-2,6-dihydroxy-2-(2-methoxy-2-oxoethyl)-6-methylheptanoyl]cephalotaxine, 1,4-DIAMINOBUTANE, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-15 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
8G5Y
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (IC state) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 28S rRNA, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
8G60
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (CR state) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 28S rRNA, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
8G61
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (AC state) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 28S rRNA, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
8GLP
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (Consensus LSU focused refined structure) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Watson, Z.L, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-03-22 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (1.67 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
8G5Z
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (GA state) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 28S rRNA, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
3LJN
 
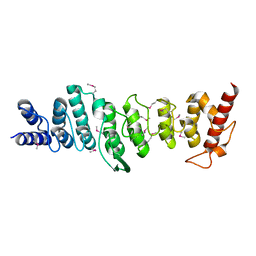 | |
3LEF
 
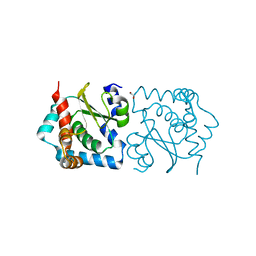 | | Crystal structure of HIV epitope-scaffold 4E10_S0_1Z6NA_001 | | Descriptor: | 1,2-ETHANEDIOL, Uncharacterized protein 4E10_S0_1Z6NA_001 (T18) | | Authors: | Holmes, M.A. | | Deposit date: | 2010-01-14 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Computational Design of Epitope-Scaffolds Allows Induction of Antibodies Specific for a Poorly Immunogenic HIV Vaccine Epitope.
Structure, 18, 2010
|
|
3H3P
 
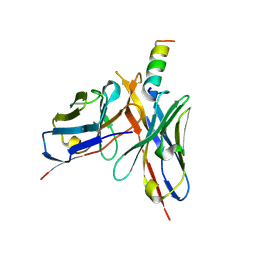 | | Crystal structure of HIV epitope-scaffold 4E10 Fv complex | | Descriptor: | 4E10_S0_1TJLC_004_N, CALCIUM ION, Fv 4E10 heavy chain, ... | | Authors: | Holmes, M.A. | | Deposit date: | 2009-04-16 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Interactions between lipids and human anti-HIV antibody 4E10 can be reduced without ablating neutralizing activity
J.Virol., 84, 2010
|
|
1BVK
 
 | |
3T43
 
 | |
3LHP
 
 | |
3LH2
 
 | |
1BVL
 
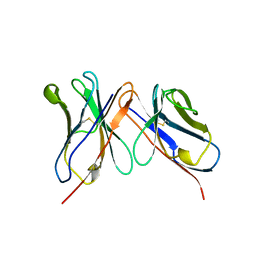 | | HUMANIZED ANTI-LYSOZYME FV | | Descriptor: | HULYS11 | | Authors: | Holmes, M.A, Foote, J. | | Deposit date: | 1998-09-16 | | Release date: | 1999-02-16 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structural consequences of humanizing an antibody.
J.Immunol., 158, 1997
|
|
1X71
 
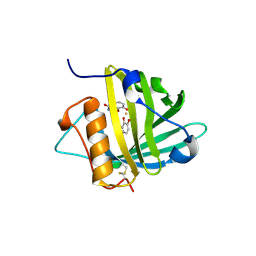 | | Crystal structure of Siderocalin (NGAL, Lipocalin 2) complexed with TRENCAM-3,2-HOPO, a cepabactin analogue | | Descriptor: | 2,3-DIHYDROXYBENZAMIDE, FE (III) ION, Neutrophil gelatinase-associated lipocalin | | Authors: | Holmes, M.A, Paulsene, W, Jide, X, Ratledge, C, Strong, R.K. | | Deposit date: | 2004-08-12 | | Release date: | 2005-01-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Siderocalin (Lcn 2) Also Binds Carboxymycobactins, Potentially Defending against Mycobacterial Infections through Iron Sequestration
Structure, 13, 2005
|
|
3LG7
 
 | | Crystal structure of HIV epitope-scaffold 4E10_S0_1EZ3A_002_C | | Descriptor: | 4E10_S0_1EZ3A_002_C (T246), SULFATE ION | | Authors: | Holmes, M.A. | | Deposit date: | 2010-01-19 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Computational Design of Epitope-Scaffolds Allows Induction of Antibodies Specific for a Poorly Immunogenic HIV Vaccine Epitope.
Structure, 18, 2010
|
|
1X8U
 
 | | Crystal structure of Siderocalin (NGAL, Lipocalin 2) complexed with Carboxymycobactin T | | Descriptor: | CARBOXYMYCOBACTIN T, Neutrophil gelatinase-associated lipocalin | | Authors: | Holmes, M.A, Paulsene, W, Jide, X, Ratledge, C, Strong, R.K. | | Deposit date: | 2004-08-18 | | Release date: | 2005-01-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Siderocalin (Lcn 2) Also Binds Carboxymycobactins, Potentially Defending against Mycobacterial Infections through Iron Sequestration
Structure, 13, 2005
|
|
1X89
 
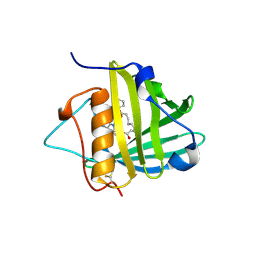 | | Crystal structure of Siderocalin (NGAL, Lipocalin 2) complexed with Carboxymycobactin S | | Descriptor: | CARBOXYMYCOBACTIN S, Neutrophil gelatinase-associated lipocalin | | Authors: | Holmes, M.A, Paulsene, W, Jide, X, Ratledge, C, Strong, R.K. | | Deposit date: | 2004-08-17 | | Release date: | 2005-01-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Siderocalin (Lcn 2) Also Binds Carboxymycobactins, Potentially Defending against Mycobacterial Infections through Iron Sequestration
Structure, 13, 2005
|
|
2HMQ
 
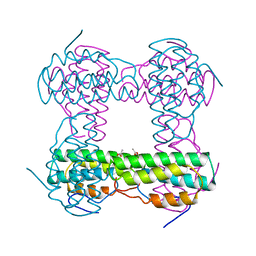 | |
2HMZ
 
 | |
1HMD
 
 | | THE STRUCTURE OF DEOXY AND OXY HEMERYTHRIN AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | ACETYL GROUP, HEMERYTHRIN, MU-OXO-DIIRON | | Authors: | Holmes, M.A, Letrong, I, Turley, S, Sieker, L.C, Stenkamp, R.E. | | Deposit date: | 1990-10-18 | | Release date: | 1992-01-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of deoxy and oxy hemerythrin at 2.0 A resolution.
J.Mol.Biol., 218, 1991
|
|
1HMO
 
 | | THE STRUCTURE OF DEOXY AND OXY HEMERYTHRIN AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | ACETYL GROUP, HEMERYTHRIN, MU-OXO-DIIRON, ... | | Authors: | Holmes, M.A, Letrong, I, Turley, S, Sieker, L.C, Stenkamp, R.E. | | Deposit date: | 1990-10-18 | | Release date: | 1992-01-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of deoxy and oxy hemerythrin at 2.0 A resolution.
J.Mol.Biol., 218, 1991
|
|
2A03
 
 | |
2B4W
 
 | |
