5FEU
 
 | | Noroxomaritidine/Norcraugsodine Reductase in complex with NADP+ | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Noroxomaritidine/Norcraugsodine Reductase | | Authors: | Holland, C, Jez, J.M. | | Deposit date: | 2015-12-17 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Identification of a Noroxomaritidine Reductase with Amaryllidaceae Alkaloid Biosynthesis Related Activities.
J.Biol.Chem., 291, 2016
|
|
4GKZ
 
 | | HA1.7, a MHC class II restricted TCR specific for haemagglutinin | | Descriptor: | 1,2-ETHANEDIOL, Alpha chain of Class II TCR, Beta Chain of Class II TCR, ... | | Authors: | Holland, C.J, Rizkallah, P.J, Cole, D.K, Sewell, A.K, Godkin, A.J. | | Deposit date: | 2012-08-13 | | Release date: | 2012-11-07 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Minimal conformational plasticity enables TCR cross-reactivity to different MHC class II heterodimers.
Sci Rep, 2, 2012
|
|
5T95
 
 | | Prephenate Dehydrogenase M219T, N222D mutant from Soybean | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Prephenate dehydrogenase 1, TYROSINE | | Authors: | Holland, C.K, Jez, J.M. | | Deposit date: | 2016-09-09 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.689 Å) | | Cite: | Molecular basis of the evolution of alternative tyrosine biosynthetic routes in plants.
Nat. Chem. Biol., 13, 2017
|
|
5T9E
 
 | |
5T9F
 
 | | Prephenate Dehydrogenase N222D mutant from Soybean | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Prephenate dehydrogenase 1, TYROSINE | | Authors: | Holland, C.K, Jez, J.M. | | Deposit date: | 2016-09-09 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | Molecular basis of the evolution of alternative tyrosine biosynthetic routes in plants.
Nat. Chem. Biol., 13, 2017
|
|
5WMH
 
 | | Arabidopsis thaliana prephenate aminotransferase | | Descriptor: | Bifunctional aspartate aminotransferase and glutamate/aspartate-prephenate aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Holland, C.K, Jez, J.M. | | Deposit date: | 2017-07-28 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for substrate recognition and inhibition of prephenate aminotransferase from Arabidopsis.
Plant J., 94, 2018
|
|
5W6Y
 
 | | Physcomitrella patens Chorismate Mutase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Chorismate mutase, TRYPTOPHAN | | Authors: | Holland, C.K, Kroll, K, Jez, J.M. | | Deposit date: | 2017-06-18 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Evolution of allosteric regulation in chorismate mutases from early plants.
Biochem. J., 474, 2017
|
|
5WHX
 
 | | PREPHENATE DEHYDROGENASE FROM SOYBEAN | | Descriptor: | CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Prephenate dehydrogenase 1 | | Authors: | Holland, C.K, Jez, J.M. | | Deposit date: | 2017-07-18 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Molecular basis of the evolution of alternative tyrosine biosynthetic routes in plants.
Nat. Chem. Biol., 13, 2017
|
|
6OMS
 
 | | Arabidopsis GH3.12 with Chorismate | | Descriptor: | (3R,4R)-3-[(1-carboxyethenyl)oxy]-4-hydroxycyclohexa-1,5-diene-1-carboxylic acid, 4-substituted benzoates-glutamate ligase GH3.12, ADENOSINE MONOPHOSPHATE | | Authors: | Zubieta, C, Westfall, C.S, Holland, C.K, Jez, J.M. | | Deposit date: | 2019-04-19 | | Release date: | 2019-10-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.942 Å) | | Cite: | Brassicaceae-specific Gretchen Hagen 3 acyl acid amido synthetases conjugate amino acids to chorismate, a precursor of aromatic amino acids and salicylic acid.
J.Biol.Chem., 294, 2019
|
|
5FFF
 
 | |
5FF9
 
 | |
3BZ5
 
 | | Functional domain of InlJ from Listeria monocytogenes includes a cysteine ladder | | Descriptor: | CHLORIDE ION, Internalin-J, SULFATE ION | | Authors: | Bublitz, M, Holland, C, Sabet, C, Reichelt, J, Cossart, P, Heinz, D.W, Bierne, H, Schubert, W.D. | | Deposit date: | 2008-01-17 | | Release date: | 2008-06-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and standardized geometric analysis of InlJ, a listerial virulence factor and leucine-rich repeat protein with a novel cysteine ladder.
J.Mol.Biol., 378, 2008
|
|
5WML
 
 | | Arabidopsis thaliana Prephenate Aminotransferase mutant- K306A | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Bifunctional aspartate aminotransferase and glutamate/aspartate-prephenate aminotransferase, GLUTAMIC ACID | | Authors: | Jez, J.M, Holland, C.K. | | Deposit date: | 2017-07-29 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Structural basis for substrate recognition and inhibition of prephenate aminotransferase from Arabidopsis.
Plant J., 94, 2018
|
|
5WMK
 
 | |
5WMI
 
 | | Arabidopsis thaliana Prephenate Aminotransferase mutant- T84V | | Descriptor: | 2-OXOGLUTARIC ACID, Bifunctional aspartate aminotransferase and glutamate/aspartate-prephenate aminotransferase | | Authors: | Jez, J.M, Holland, C.K. | | Deposit date: | 2017-07-28 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for substrate recognition and inhibition of prephenate aminotransferase from Arabidopsis.
Plant J., 94, 2018
|
|
3FI7
 
 | | Crystal Structure of the autolysin Auto (Lmo1076) from Listeria monocytogenes, catalytic domain | | Descriptor: | Lmo1076 protein, SULFATE ION | | Authors: | Bublitz, M, Polle, L, Holland, C, Nimtz, M, Heinz, D.W, Schubert, W.D. | | Deposit date: | 2008-12-11 | | Release date: | 2009-04-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for autoinhibition and activation of Auto, a virulence-associated peptidoglycan hydrolase of Listeria monocytogenes.
Mol.Microbiol., 71, 2009
|
|
8WKY
 
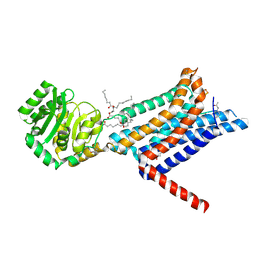 | | Crystal structure of the Melanocortin-4 Receptor (MC4R) in complex with S25 | | Descriptor: | CALCIUM ION, Melanocortin receptor 4, N-(2-aminoethyl)-5-(2-{[4-(morpholin-4-yl)pyridin-2-yl]amino}-1,3-thiazol-5-yl)pyridine-3-carboxamide, ... | | Authors: | Gimenez, L.E, Martin, C, Yu, J, Hollanders, C, Hernandez, C, Dahir, N.S, Wu, Y, Yao, D, Han, G.W, Wu, L, Poorten, O.V, Lamouroux, A, Mannes, M, Tourwe, D, Zhao, S, Stevens, R.C, Cone, R.D, Ballet, S. | | Deposit date: | 2023-09-28 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Novel Cocrystal Structures of Peptide Antagonists Bound to the Human Melanocortin Receptor 4 Unveil Unexplored Grounds for Structure-Based Drug Design.
J.Med.Chem., 67, 2024
|
|
8WKZ
 
 | | Crystal structure of the Melanocortin-4 Receptor (MC4R) in complex with S31 | | Descriptor: | CALCIUM ION, Melanocortin receptor 4, OLEIC ACID, ... | | Authors: | Gimenez, L.E, Martin, C, Yu, J, Hollanders, C, Hernandez, C, Dahir, N.S, Wu, Y, Yao, D, Han, G.W, Wu, L, Poorten, O.V, Lamouroux, A, Mannes, M, Tourwe, D, Zhao, S, Stevens, R.C, Cone, R.D, Ballet, S. | | Deposit date: | 2023-09-28 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Novel Cocrystal Structures of Peptide Antagonists Bound to the Human Melanocortin Receptor 4 Unveil Unexplored Grounds for Structure-Based Drug Design.
J.Med.Chem., 67, 2024
|
|
