5IEI
 
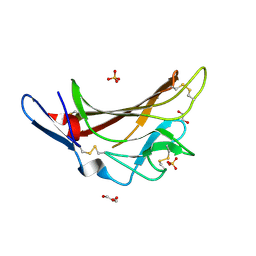 | | X-ray crystallographic structure of a high affinity IGF2 antagonist (Domain11 AB5 RHH) based on human IGF2R domain 11 | | Descriptor: | 1,2-ETHANEDIOL, Cation-independent mannose-6-phosphate receptor, GLYCEROL, ... | | Authors: | Nicholls, R.D, Williams, C, Strickland, M, Frago, S, Hassan, A.B, Crump, M.P. | | Deposit date: | 2016-02-25 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Functional evolution of IGF2:IGF2R domain 11 binding generates novel structural interactions and a specific IGF2 antagonist.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3MLE
 
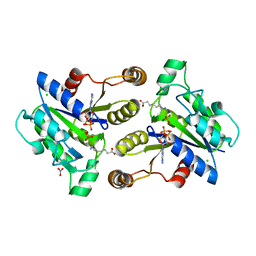 | | Crystal structure of dethiobiotin synthetase (BioD) from Helicobacter pylori cocrystallized with ATP | | Descriptor: | 8-aminooctanoic acid, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Nicholls, R, Porebski, P.J, Klimecka, M.M, Chruszcz, M, Murzyn, K, Joachimiak, A, Murshudov, G, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-16 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural characterization of Helicobacter pylori dethiobiotin synthetase reveals differences between family members.
Febs J., 279, 2012
|
|
4ND9
 
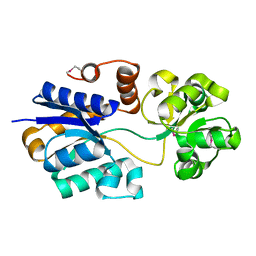 | | The putative substrate binding domain of ABC-type transporter from Agrobacterium tumefaciens in open conformation | | Descriptor: | ABC transporter, substrate binding protein (Proline/glycine/betaine) | | Authors: | Nicholls, R, Tkaczuk, K.L, Kagan, O, Chruszcz, M, Domagalski, M.J, Savchenko, A, Joachimiak, A, Murshudov, G, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-10-25 | | Release date: | 2013-12-11 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The putative substrate binding domain of ABC-type transporter from Agrobacterium tumefaciens in open conformation
To be Published
|
|
3PDS
 
 | | Irreversible Agonist-Beta2 Adrenoceptor Complex | | Descriptor: | 8-hydroxy-5-[(1R)-1-hydroxy-2-({2-[3-methoxy-4-(3-sulfanylpropoxy)phenyl]ethyl}amino)ethyl]quinolin-2(1H)-one, CHOLESTEROL, Fusion protein Beta-2 adrenergic receptor/Lysozyme, ... | | Authors: | Rosenbaum, D.M, Zhang, C, Lyons, J.A, Holl, R, Aragao, D, Arlow, D.H, Rasmussen, S.G.F, Choi, H.-J, DeVree, B.T, Sunahara, R.K, Chae, P.S, Gellman, S.H, Dror, R.O, Shaw, D.E, Weis, W.I, Caffrey, M, Gmeiner, P, Kobilka, B.K. | | Deposit date: | 2010-10-24 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure and function of an irreversible agonist-beta(2) adrenoceptor complex
Nature, 469, 2011
|
|
4ROT
 
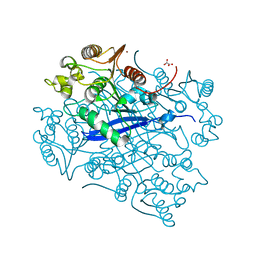 | | Crystal structure of esterase A from Streptococcus pyogenes | | Descriptor: | Esterase A, OXALATE ION, ZINC ION | | Authors: | Bennett, M.D, Holland, R, Coulibaly, F, Loo, T.S, Norris, G.E, Anderson, B.F. | | Deposit date: | 2014-10-28 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of esterase A from Streptococcus pyogenes
To be Published
|
|
6Z32
 
 | | Human cation-independent mannose 6-phosphate/IGF2 receptor domains 7-11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cation-independent mannose-6-phosphate receptor, SULFATE ION, ... | | Authors: | Bochel, A.J, Williams, C, McCoy, A.J, Hoppe, H, Winter, A.J, Nicholls, R.D, Harlos, K, Jones, Y.E, Berger, I, Hassan, B, Crump, M.P. | | Deposit date: | 2020-05-19 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | Structure of the Human Cation-Independent Mannose 6-Phosphate/IGF2 Receptor Domains 7-11 Uncovers the Mannose 6-Phosphate Binding Site of Domain 9.
Structure, 28, 2020
|
|
4IUW
 
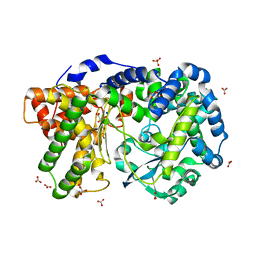 | | Crystal structure of PEPO from Lactobacillus rhamnosis HN001 (DR20) | | Descriptor: | CARBONATE ION, CITRIC ACID, Neutral endopeptidase, ... | | Authors: | Anderson, B.F, Knapp, K.M, Holland, R, Norris, G.E, Christensson, C, Bratt, H, Collins, L.J, Coolbear, T, Lubbers, M.W, Toole, P.W.O, Reid, J.R, Jameson, G.B. | | Deposit date: | 2013-01-21 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of PEPO from Lactobacillus rhamnosis HN001 (DR20)
TO BE PUBLISHED
|
|
5AKB
 
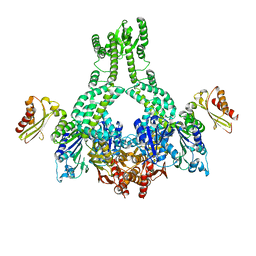 | | MutS in complex with the N-terminal domain of MutL - crystal form 1 | | Descriptor: | DNA MISMATCH REPAIR PROTEIN MUTL, DNA MISMATCH REPAIR PROTEIN MUTS, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Groothuizen, F.S, Winkler, I, Cristovao, M, Fish, A, Winterwerp, H.H.K, Reumer, A, Marx, A.D, Hermans, N, Nicholls, R.A, Murshudov, G.N, Lebbink, J.H.G, Friedhoff, P, Sixma, T.K. | | Deposit date: | 2015-03-03 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.71 Å) | | Cite: | MutS/MutL crystal structure reveals that the MutS sliding clamp loads MutL onto DNA.
Elife, 4, 2015
|
|
5AKC
 
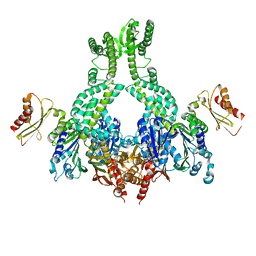 | | MutS in complex with the N-terminal domain of MutL - crystal form 2 | | Descriptor: | DNA MISMATCH REPAIR PROTEIN MUTL, DNA MISMATCH REPAIR PROTEIN MUTS, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Groothuizen, F.S, Winkler, I, Cristovao, M, Fish, A, Winterwerp, H.H.K, Reumer, A, Marx, A.D, Hermans, N, Nicholls, R.A, Murshudov, G.N, Lebbink, J.H.G, Friedhoff, P, Sixma, T.K. | | Deposit date: | 2015-03-03 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (6.6 Å) | | Cite: | MutS/MutL crystal structure reveals that the MutS sliding clamp loads MutL onto DNA.
Elife, 4, 2015
|
|
5AKD
 
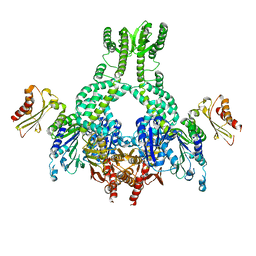 | | MutS in complex with the N-terminal domain of MutL - crystal form 3 | | Descriptor: | DNA MISMATCH REPAIR PROTEIN MUTL, DNA MISMATCH REPAIR PROTEIN MUTS, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Groothuizen, F.S, Winkler, I, Cristovao, M, Fish, A, Winterwerp, H.H.K, Reumer, A, Marx, A.D, Hermans, N, Nicholls, R.A, Murshudov, G.N, Lebbink, J.H.G, Friedhoff, P, Sixma, T.K. | | Deposit date: | 2015-03-03 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (7.6 Å) | | Cite: | MutS/MutL crystal structure reveals that the MutS sliding clamp loads MutL onto DNA.
Elife, 4, 2015
|
|
4N5H
 
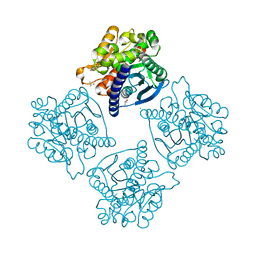 | | Crystal structure of ESTERASE B from Lactobacillus Rhamnosis (HN001) | | Descriptor: | CALCIUM ION, CHLORIDE ION, Esterase/lipase, ... | | Authors: | Bennett, M.D, Holland, R, Loo, T.S, Smith, C.A, Norris, G.E, Delabre, M.-L, Anderson, B.F. | | Deposit date: | 2013-10-09 | | Release date: | 2014-10-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal structure of ESTERASE B from Lactobacillus Rhamnosis (HN001)
TO BE PUBLISHED
|
|
4OUK
 
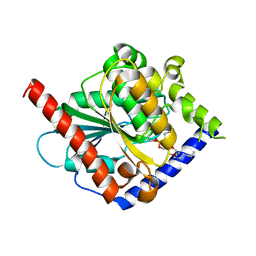 | | Crystal structure of a C6-C4 SN3 inhibited ESTERASE B from LACTOBACILLUS RHAMNOSIS | | Descriptor: | (2R)-2,3-bis(hexyloxy)propyl hydrogen (S)-pentylphosphonate, Esterase B | | Authors: | Colbert, D.A, Bennett, M.D, Lun, D.J, Holland, R, Delabre, M.-L, Loo, T.S, Anderson, B.F, Norris, G.E. | | Deposit date: | 2014-02-17 | | Release date: | 2014-08-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a C6-C4 SN3 inhibited ESTERASE B from LACTOBACILLUS RHAMNOSIS
TO BE PUBLISHED
|
|
4NE4
 
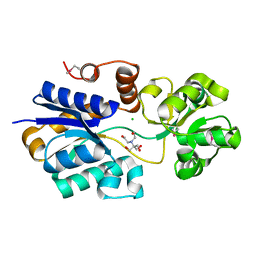 | | Crystal structure of ABC transporter substrate binding protein ProX from Agrobacterium tumefaciens cocrystalized with BTB | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ABC transporter, substrate binding protein (Proline/glycine/betaine), ... | | Authors: | Tkaczuk, K.L, Nicholls, R, Kagan, O, Chruszcz, M, Domagalski, M.J, Savchenko, A, Joachimiak, A, Murshudov, G, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-10-28 | | Release date: | 2013-11-27 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of ABC transporter substrate binding protein ProX from Agrobacterium tumefaciens cocrystalized with BTB
To be Published
|
|
3E1G
 
 | |
3DYV
 
 | |
3DLT
 
 | |
3DKR
 
 | |
3DYI
 
 | |
