1AR7
 
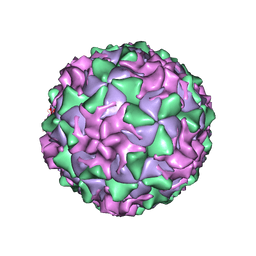 | | P1/MAHONEY POLIOVIRUS, DOUBLE MUTANT P1095S + H2142Y | | Descriptor: | MYRISTIC ACID, P1/MAHONEY POLIOVIRUS, SPHINGOSINE | | Authors: | Wien, M.W, Curry, S, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-08-11 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of poliovirus mutants that overcome receptor defects.
Nat.Struct.Biol., 4, 1997
|
|
1T6L
 
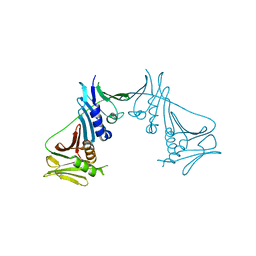 | | Crystal Structure of the Human Cytomegalovirus DNA Polymerase Subunit, UL44 | | Descriptor: | DNA polymerase processivity factor | | Authors: | Appleton, B.A, Loregian, A, Filman, D.J, Coen, D.M, Hogle, J.M. | | Deposit date: | 2004-05-06 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Cytomegalovirus DNA Polymerase Subunit UL44 Forms a C Clamp-Shaped Dimer.
Mol.Cell, 15, 2004
|
|
1SEL
 
 | |
3ZX8
 
 | | Cryo-EM reconstruction of native and expanded Turnip Crinkle virus | | Descriptor: | CAPSID PROTEIN | | Authors: | Bakker, S.E, Robottom, J, Hogle, J.M, Maeda, A, Pearson, A.R, Stockley, P.G, Ranson, N.A, Harrison, S.C. | | Deposit date: | 2011-08-08 | | Release date: | 2012-07-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Isolation of an Asymmetric RNA Uncoating Intermediate for a Single-Stranded RNA Plant Virus.
J.Mol.Biol., 417, 2012
|
|
3I2M
 
 | | The Crystal Structure of PF-8, the DNA Polymerase Accessory Subunit from Kaposi s Sarcoma-Associated Herpesvirus | | Descriptor: | ORF59 | | Authors: | Baltz, J.L, Filman, D.J, Ciustea, M, Silverman, J.E.Y, Lautenschlager, C.L, Coen, D.M, Ricciardi, R.P, Hogle, J.M. | | Deposit date: | 2009-06-29 | | Release date: | 2010-05-12 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The crystal structure of PF-8, the DNA polymerase accessory subunit from Kaposi's sarcoma-associated herpesvirus.
J.Virol., 83, 2009
|
|
3IYB
 
 | | Poliovirus early RNA-release intermediate | | Descriptor: | Genome polyprotein, Precursor polyprotein, VP1 core | | Authors: | Levy, H.C, Bostina, M, Filman, D.J, Hogle, J.M. | | Deposit date: | 2009-07-21 | | Release date: | 2010-03-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Catching a virus in the act of RNA release: a novel poliovirus uncoating intermediate characterized by cryo-electron microscopy.
J.Virol., 84, 2010
|
|
3JBE
 
 | | Complex of poliovirus with VHH PVSS8A | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Strauss, M, Schotte, L, Thys, B, Filman, D.J, Hogle, J.M. | | Deposit date: | 2015-08-26 | | Release date: | 2016-01-27 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Five of Five VHHs Neutralizing Poliovirus Bind the Receptor-Binding Site.
J.Virol., 90, 2016
|
|
3J3P
 
 | | Conformational Shift of a Major Poliovirus Antigen Confirmed by Immuno-Cryogenic Electron Microscopy: 135S Poliovirus and C3-Fab Complex | | Descriptor: | C3 antibody, heavy chain, light chain, ... | | Authors: | Lin, J, Cheng, N, Hogle, J.M, Steven, A.C, Belnap, D.M. | | Deposit date: | 2013-04-10 | | Release date: | 2013-07-03 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | Conformational shift of a major poliovirus antigen confirmed by immuno-cryogenic electron microscopy.
J.Immunol., 191, 2013
|
|
3J48
 
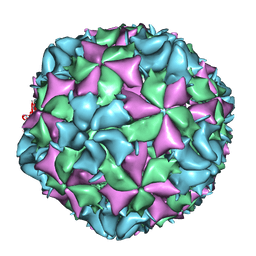 | | Cryo-EM structure of Poliovirus 135S particles | | Descriptor: | Protein VP1, Protein VP2, Protein VP3 | | Authors: | Butan, C, Fiman, D.J, Hogle, J.M. | | Deposit date: | 2013-06-28 | | Release date: | 2013-12-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Cryo-Electron Microscopy Reconstruction Shows Poliovirus 135S Particles Poised for Membrane Interaction and RNA Release.
J.Virol., 88, 2014
|
|
3J8F
 
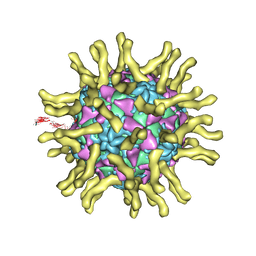 | | Cryo-EM reconstruction of poliovirus-receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid protein VP1, ... | | Authors: | Strauss, M, Filman, D.J, Belnap, D.M, Cheng, N, Noel, R.T, Hogle, J.M. | | Deposit date: | 2014-10-20 | | Release date: | 2015-02-11 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Nectin-Like Interactions between Poliovirus and Its Receptor Trigger Conformational Changes Associated with Cell Entry.
J.Virol., 89, 2015
|
|
3J3O
 
 | | Conformational Shift of a Major Poliovirus Antigen Confirmed by Immuno-Cryogenic Electron Microscopy: 160S Poliovirus and C3-Fab Complex | | Descriptor: | C3 antibody, heavy chain, light chain, ... | | Authors: | Lin, J, Cheng, N, Hogle, J.M, Steven, A.C, Belnap, D.M. | | Deposit date: | 2013-04-10 | | Release date: | 2013-07-03 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11.1 Å) | | Cite: | Conformational shift of a major poliovirus antigen confirmed by immuno-cryogenic electron microscopy.
J.Immunol., 191, 2013
|
|
3JBF
 
 | | Complex of poliovirus with VHH PVSP19B | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Strauss, M, Schotte, L, Thys, B, Filman, D.J, Hogle, J.M. | | Deposit date: | 2015-08-26 | | Release date: | 2016-01-27 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Five of Five VHHs Neutralizing Poliovirus Bind the Receptor-Binding Site.
J.Virol., 90, 2016
|
|
3JBD
 
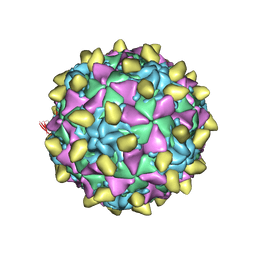 | | Complex of poliovirus with VHH PVSP6A | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Strauss, M, Schotte, L, Thys, B, Filman, D.J, Hogle, J.M. | | Deposit date: | 2015-08-26 | | Release date: | 2016-01-27 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Five of Five VHHs Neutralizing Poliovirus Bind the Receptor-Binding Site.
J.Virol., 90, 2016
|
|
3JBC
 
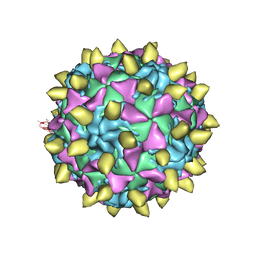 | | Complex of Poliovirus with VHH PVSP29F | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Strauss, M, Schotte, L, Thys, B, Filman, D.J, Hogle, J.M. | | Deposit date: | 2015-08-26 | | Release date: | 2016-01-27 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Five of Five VHHs Neutralizing Poliovirus Bind the Receptor-Binding Site.
J.Virol., 90, 2016
|
|
3JBG
 
 | | Complex of poliovirus with VHH PVSS21E | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Strauss, M, Schotte, L, Thys, B, Filman, D.J, Hogle, J.M. | | Deposit date: | 2015-08-26 | | Release date: | 2016-01-27 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Five of Five VHHs Neutralizing Poliovirus Bind the Receptor-Binding Site.
J.Virol., 90, 2016
|
|
3IYC
 
 | | Poliovirus late RNA-release intermediate | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Genome polyprotein, ... | | Authors: | Levy, H.C, Bostina, M, Filman, D.J, Hogle, J.M. | | Deposit date: | 2009-07-21 | | Release date: | 2010-03-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Catching a virus in the act of RNA release: a novel poliovirus uncoating intermediate characterized by cryo-electron microscopy.
J.Virol., 84, 2010
|
|
3J9F
 
 | | Poliovirus complexed with soluble, deglycosylated poliovirus receptor (Pvr) at 4 degrees C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITIC ACID, ... | | Authors: | Strauss, M, Filman, D.J, Belnap, D.M, Cheng, N, Noel, R.T, Hogle, J.M. | | Deposit date: | 2015-01-15 | | Release date: | 2015-02-11 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Nectin-Like Interactions between Poliovirus and Its Receptor Trigger Conformational Changes Associated with Cell Entry.
J.Virol., 89, 2015
|
|
3HSL
 
 | | The Crystal Structure of PF-8, the DNA Polymerase Accessory Subunit from Kaposi's Sarcoma-Associated Herpesvirus | | Descriptor: | ORF59 | | Authors: | Baltz, J.L, Filman, D.J, Ciustea, M, Silverman, J.E.Y, Lautenschlager, C.L, Coen, D.M, Ricciardi, R.P, Hogle, J.M. | | Deposit date: | 2009-06-10 | | Release date: | 2009-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of PF-8, the DNA polymerase accessory subunit from Kaposi's sarcoma-associated herpesvirus.
J.Virol., 83, 2009
|
|
1DML
 
 | | CRYSTAL STRUCTURE OF HERPES SIMPLEX UL42 BOUND TO THE C-TERMINUS OF HSV POL | | Descriptor: | DNA POLYMERASE, DNA POLYMERASE PROCESSIVITY FACTOR | | Authors: | Zuccola, H.J, Filman, D.J, Coen, D.M, Hogle, J.M. | | Deposit date: | 1999-12-14 | | Release date: | 2000-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of an unusual processivity factor, herpes simplex virus UL42, bound to the C terminus of its cognate polymerase.
Mol.Cell, 5, 2000
|
|
1DPS
 
 | | THE CRYSTAL STRUCTURE OF DPS, A FERRITIN HOMOLOG THAT BINDS AND PROTECTS DNA | | Descriptor: | DPS, SODIUM ION | | Authors: | Grant, R.A, Filman, D.J, Finkel, S.E, Kolter, R, Hogle, J.M. | | Deposit date: | 1998-02-23 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of Dps, a ferritin homolog that binds and protects DNA.
Nat.Struct.Biol., 5, 1998
|
|
1A92
 
 | |
