2JVN
 
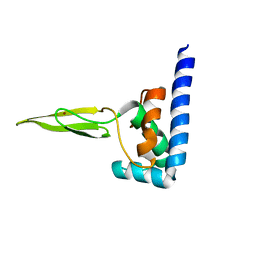 | | Domain C of human PARP-1 | | Descriptor: | Poly [ADP-ribose] polymerase 1, ZINC ION | | Authors: | Hoffman, D, Tao, Z, Liu, H. | | Deposit date: | 2007-09-24 | | Release date: | 2008-05-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Domain C of human poly(ADP-ribose) polymerase-1 is important for enzyme activity and contains a novel zinc-ribbon motif.
Biochemistry, 47, 2008
|
|
6Y98
 
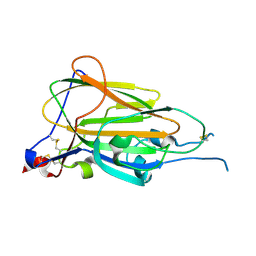 | | Crystal Structure of subtype-switched Epithelial Adhesin 9 to 1 A domain (Epa9-CBL2Epa1) from Candida glabrata in complex with beta-lactose | | Descriptor: | CALCIUM ION, PA14 domain-containing protein, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Hoffmann, D, Diderrich, R, Kock, M, Friederichs, S, Reithofer, V, Essen, L.-O, Moesch, H.-U. | | Deposit date: | 2020-03-06 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Functional reprogramming ofCandida glabrataepithelial adhesins: the role of conserved and variable structural motifs in ligand binding.
J.Biol.Chem., 295, 2020
|
|
6Y9J
 
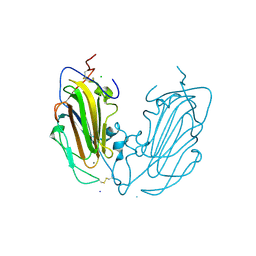 | | Crystal Structure of subtype-switched Epithelial Adhesin 1 to 9 A domain (Epa1-CBL2Epa9) from Candida glabrata in complex with beta-lactose | | Descriptor: | CALCIUM ION, CHLORIDE ION, Epa1p, ... | | Authors: | Hoffmann, D, Diderrich, R, Kock, M, Friederichs, S, Reithofer, V, Essen, L.-O, Moesch, H.-U. | | Deposit date: | 2020-03-09 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Functional reprogramming ofCandida glabrataepithelial adhesins: the role of conserved and variable structural motifs in ligand binding.
J.Biol.Chem., 295, 2020
|
|
3DDF
 
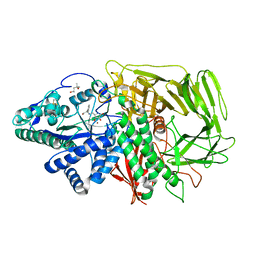 | | GOLGI MANNOSIDASE II complex with (3R,4R,5R)-3,4-Dihydroxy-5-({[(1R)-2-hydroxy-1 phenylethyl]amino}methyl) pyrrolidin-2-one | | Descriptor: | (3R,4R,5R)-3,4-dihydroxy-5-({[(1R)-2-hydroxy-1-phenylethyl]amino}methyl)pyrrolidin-2-one, (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ... | | Authors: | Kuntz, D.A, Rose, D.R, Hoffman, D. | | Deposit date: | 2008-06-05 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Functionalized pyrrolidine inhibitors of human type II alpha-mannosidases as anti-cancer agents: optimizing the fit to the active site
Bioorg.Med.Chem., 16, 2008
|
|
3DDG
 
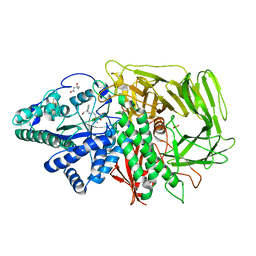 | | GOLGI MANNOSIDASE II complex with (3R,4R,5R)-3,4-Dihydroxy-5-({[(1R)-2-hydroxy-1 phenylethyl]amino}methyl) methylpyrrolidin-2-one | | Descriptor: | (3R,4R,5R)-3,4-dihydroxy-5-({[(1R)-2-hydroxy-1-phenylethyl]amino}methyl)-1-methylpyrrolidin-2-one, (4R)-2-METHYLPENTANE-2,4-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuntz, D.A, Rose, D.R, Hoffman, D. | | Deposit date: | 2008-06-05 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Functionalized pyrrolidine inhibitors of human type II alpha-mannosidases as anti-cancer agents: optimizing the fit to the active site
Bioorg.Med.Chem., 16, 2008
|
|
1CQU
 
 | |
2PPZ
 
 | |
