6OUA
 
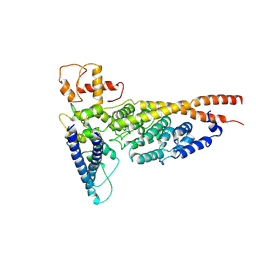 | | Cryo-EM structure of the yeast Ctf3 complex | | Descriptor: | Inner kinetochore subunit CTF3, Inner kinetochore subunit MCM16, Inner kinetochore subunit MCM22 | | Authors: | Hinshaw, S.M, Harrison, S.C. | | Deposit date: | 2019-05-04 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | The structure of the yeast Ctf3 complex.
Elife, 8, 2019
|
|
5W94
 
 | |
4XDN
 
 | | Crystal structure of Scc4 in complex with Scc2n | | Descriptor: | MAU2 chromatid cohesion factor homolog, SULFATE ION, Sister chromatid cohesion protein 2 | | Authors: | Hinshaw, S.M, Harrison, S.C. | | Deposit date: | 2014-12-19 | | Release date: | 2015-06-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural evidence for Scc4-dependent localization of cohesin loading.
Elife, 4, 2015
|
|
6WUC
 
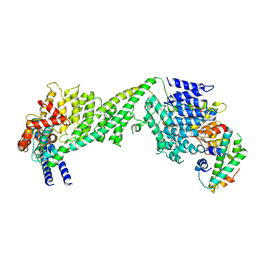 | | The yeast Ctf3 complex with Cnn1-Wip1 | | Descriptor: | Inner kinetochore subunit CNN1, Inner kinetochore subunit CTF3, Inner kinetochore subunit MCM16, ... | | Authors: | Hinshaw, S.M, Harrison, S.C. | | Deposit date: | 2020-05-04 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | The Structural Basis for Kinetochore Stabilization by Cnn1/CENP-T.
Curr.Biol., 30, 2020
|
|
6NUW
 
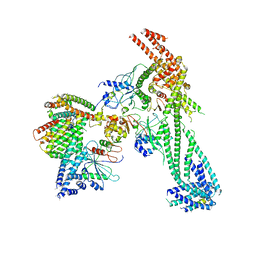 | | Yeast Ctf19 complex | | Descriptor: | Inner kinetochore subunit AME1, Inner kinetochore subunit CHL4, Inner kinetochore subunit CTF19, ... | | Authors: | Hinshaw, S.M, Harrison, S.C. | | Deposit date: | 2019-02-03 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | The structure of the Ctf19c/CCAN from budding yeast.
Elife, 8, 2019
|
|
4JE3
 
 | |
4IT3
 
 | | Crystal Structure of Iml3 from S. cerevisiae | | Descriptor: | Central kinetochore subunit IML3 | | Authors: | Hinshaw, S.M, Harrison, S.C. | | Deposit date: | 2013-01-17 | | Release date: | 2013-10-16 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | An iml3-chl4 heterodimer links the core centromere to factors required for accurate chromosome segregation.
Cell Rep, 5, 2013
|
|
7L7Q
 
 | | Ctf3c with Ulp2-KIM | | Descriptor: | Inner kinetochore subunit CTF3, Inner kinetochore subunit MCM16, Inner kinetochore subunit MCM22 | | Authors: | Hinshaw, S.M, Harrison, S.C. | | Deposit date: | 2020-12-30 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Ctf3/CENP-I provides a docking site for the desumoylase Ulp2 at the kinetochore.
J.Cell Biol., 220, 2021
|
|
7MGS
 
 | |
7MGR
 
 | |
