3O0R
 
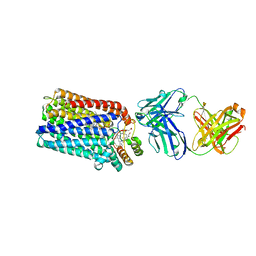 | | Crystal structure of nitric oxide reductase from Pseudomonas aeruginosa in complex with antibody fragment | | Descriptor: | CALCIUM ION, FE (III) ION, HEME C, ... | | Authors: | Hino, T, Matsumoto, Y, Nagano, S, Sugimoto, H, Fukumori, Y, Murata, T, Iwata, S, Shiro, Y. | | Deposit date: | 2010-07-20 | | Release date: | 2010-12-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of biological N2O generation by bacterial nitric oxide reductase
Science, 330, 2010
|
|
8J12
 
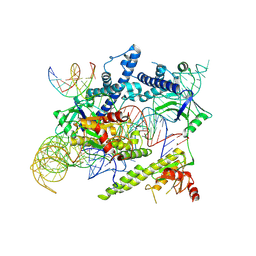 | | Cryo-EM structure of the AsCas12f-sgRNA-target DNA ternary complex | | Descriptor: | DNA (38-MER), MAGNESIUM ION, RNA (247-MER), ... | | Authors: | Hino, T, Omura, N.S, Nakagawa, R, Togashi, T, Takeda, N.S, Hiramoto, T, Tasaka, S, Hirano, H, Tokuyama, T, Uosaki, H, Ishiguro, H, Yamano, H, Ozaki, Y, Motooka, D, Mori, H, Kirita, Y, Kise, Y, Itoh, Y, Matoba, S, Aburatani, H, Yachie, N, Siksnys, V, Ohmori, T, Hoshino, A, Nureki, O. | | Deposit date: | 2023-04-12 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | An AsCas12f-based compact genome-editing tool derived by deep mutational scanning and structural analysis.
Cell, 186, 2023
|
|
8J3R
 
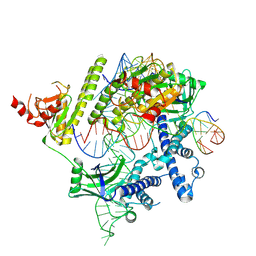 | | Cryo-EM structure of the AsCas12f-HKRA-sgRNAS3-5v7-target DNA | | Descriptor: | DNA (37-MER), DNA (38-MER), MAGNESIUM ION, ... | | Authors: | Hino, T, Omura, N.S, Nakagawa, R, Togashi, T, Takeda, N.S, Hiramoto, T, Tasaka, S, Hirano, H, Tokuyama, T, Uosaki, H, Ishiguro, H, Yamano, H, Ozaki, Y, Motooka, D, Mori, H, Kirita, Y, Kise, Y, Itoh, Y, Matoba, S, Aburatani, H, Yachie, N, Siksnys, V, Ohmori, T, Hoshino, A, Nureki, O. | | Deposit date: | 2023-04-18 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | An AsCas12f-based compact genome-editing tool derived by deep mutational scanning and structural analysis.
Cell, 186, 2023
|
|
8J1J
 
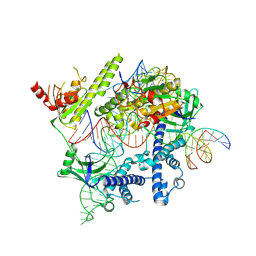 | | Cryo-EM structure of the AsCas12f-YHAM-sgRNAS3-5v7-target DNA | | Descriptor: | DNA (38-MER), MAGNESIUM ION, RNA (118-MER), ... | | Authors: | Hino, T, Omura, N.S, Nakagawa, R, Togashi, T, Takeda, N.S, Hiramoto, T, Tasaka, S, Hirano, H, Tokuyama, T, Uosaki, H, Ishiguro, H, Yamano, H, Ozaki, Y, Motooka, D, Mori, H, Kirita, Y, Kise, Y, Itoh, Y, Matoba, S, Aburatani, H, Yachie, N, Siksnys, V, Ohmori, T, Hoshino, A, Nureki, O. | | Deposit date: | 2023-04-13 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | An AsCas12f-based compact genome-editing tool derived by deep mutational scanning and structural analysis.
Cell, 186, 2023
|
|
1VCR
 
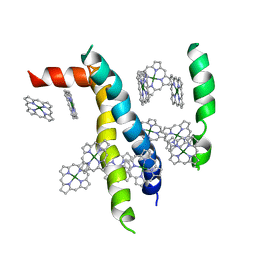 | | An icosahedral assembly of light-harvesting chlorophyll a/b protein complex from pea thylakoid membranes | | Descriptor: | CHLOROPHYLL A, CHLOROPHYLL B, Chlorophyll a-b binding protein AB80 | | Authors: | Hino, T, Kanamori, E, Shen, J.-R, Kouyama, T. | | Deposit date: | 2004-03-10 | | Release date: | 2004-03-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (9.5 Å) | | Cite: | An icosahedral assembly of the light-harvesting chlorophyll a/b protein complex from pea chloroplast thylakoid membranes.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
4Y4S
 
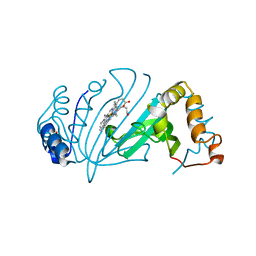 | | Crystal Structure of Y75A HasA dimer from Yersinia pseudotuberculosis | | Descriptor: | Extracellular heme acquisition hemophore HasA, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Hino, T, Kanadani, M, Muroki, T, Ishimaru, Y, Wada, Y, Sato, T, Ozaki, S. | | Deposit date: | 2015-02-11 | | Release date: | 2015-08-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of heme acquisition system A from Yersinia pseudotuberculosis (HasAypt): Roles of the axial ligand Tyr75 and two distal arginines in heme binding
J.Inorg.Biochem., 151, 2015
|
|
3VG9
 
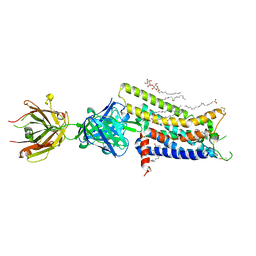 | | Crystal structure of human adenosine A2A receptor with an allosteric inverse-agonist antibody at 2.7 A resolution | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Hino, T, Arakawa, T, Iwanari, H, Yurugi-Kobayashi, T, Ikeda-Suno, C, Nakada-Nakura, Y, Kusano-Arai, O, Weyand, S, Shimamura, T, Nomura, N, Cameron, A.D, Kobayashi, T, Hamakubo, T, Iwata, S, Murata, T. | | Deposit date: | 2011-08-04 | | Release date: | 2012-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | G-protein-coupled receptor inactivation by an allosteric inverse-agonist antibody
Nature, 482, 2012
|
|
3VGA
 
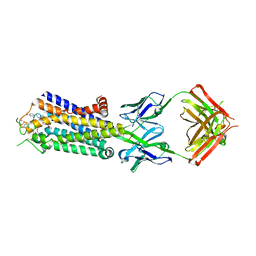 | | Crystal structure of human adenosine A2A receptor with an allosteric inverse-agonist antibody at 3.1 A resolution | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a, antibody fab fragment heavy chain, ... | | Authors: | Hino, T, Arakawa, T, Iwanari, H, Yurugi-Kobayashi, T, Ikeda-Suno, C, Nakada-Nakura, Y, Kusano-Arai, O, Weyand, S, Shimamura, T, Nomura, N, Cameron, A.D, Kobayashi, T, Hamakubo, T, Iwata, S, Murata, T. | | Deposit date: | 2011-08-04 | | Release date: | 2012-02-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | G-protein-coupled receptor inactivation by an allosteric inverse-agonist antibody
Nature, 482, 2012
|
|
5XDG
 
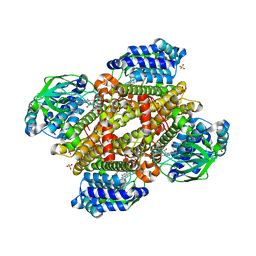 | | Crystal structure of tertiary complex of TdsC from Paenibacillus sp. A11-2 with FMN and dibenzothiophene sulfoxide | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Hino, T, Hamamoto, H, Ohshiro, T, Nagano, S. | | Deposit date: | 2017-03-28 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.747 Å) | | Cite: | Crystal structures of TdsC, a dibenzothiophene monooxygenase from the thermophile Paenibacillus sp. A11-2, reveal potential for expanding its substrate selectivity.
J. Biol. Chem., 292, 2017
|
|
5XDE
 
 | | Crystal structure of tertiary complex of TdsC from Paenibacillus sp. A11-2 with FMN and dibenzothiophene | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Hino, T, Hamamoto, H, Ohshiro, T, Nagano, S. | | Deposit date: | 2017-03-28 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of TdsC, a dibenzothiophene monooxygenase from the thermophile Paenibacillus sp. A11-2, reveal potential for expanding its substrate selectivity.
J. Biol. Chem., 292, 2017
|
|
5XDD
 
 | | Crystal structure of tertiary complex of TdsC from Paenibacillus sp. A11-2 with FMN and Indole | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, INDOLE, ... | | Authors: | Hino, T, Hamamoto, H, Ohshiro, T, Nagano, S. | | Deposit date: | 2017-03-28 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of TdsC, a dibenzothiophene monooxygenase from the thermophile Paenibacillus sp. A11-2, reveal potential for expanding its substrate selectivity.
J. Biol. Chem., 292, 2017
|
|
5XDC
 
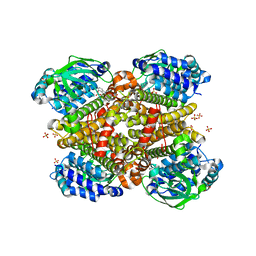 | | Crystal structure of Indole-bound TdsC from Paenibacillus sp. A11-2 | | Descriptor: | GLYCEROL, INDOLE, SULFATE ION, ... | | Authors: | Hino, T, Hamamoto, H, Ohshiro, T, Nagano, S. | | Deposit date: | 2017-03-28 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5785 Å) | | Cite: | Crystal structures of TdsC, a dibenzothiophene monooxygenase from the thermophile Paenibacillus sp. A11-2, reveal potential for expanding its substrate selectivity.
J. Biol. Chem., 292, 2017
|
|
5XB8
 
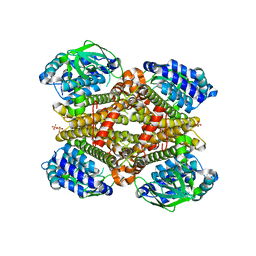 | | Crystal structure of dibenzothiophene monooxygenase (TdsC) from Paenibacillus sp. A11-2 | | Descriptor: | SULFATE ION, Thermophilic dibenzothiophene desulfurization enzyme C | | Authors: | Hino, T, Hamamoto, H, Ohshiro, T, Nagano, S. | | Deposit date: | 2017-03-16 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | Crystal structures of TdsC, a dibenzothiophene monooxygenase from the thermophile Paenibacillus sp. A11-2, reveal potential for expanding its substrate selectivity.
J. Biol. Chem., 292, 2017
|
|
5XDB
 
 | | Crystal structure of FMN-bound TdsC from Paenibacillus sp. A11-2 | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Hino, T, Hamamoto, H, Ohshiro, T, Nagano, S. | | Deposit date: | 2017-03-28 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.811 Å) | | Cite: | Crystal structures of TdsC, a dibenzothiophene monooxygenase from the thermophile Paenibacillus sp. A11-2, reveal potential for expanding its substrate selectivity.
J. Biol. Chem., 292, 2017
|
|
1HJP
 
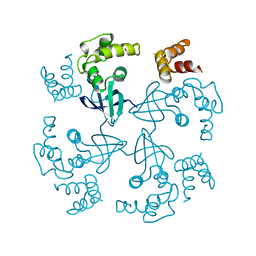 | | HOLLIDAY JUNCTION BINDING PROTEIN RUVA FROM E. COLI | | Descriptor: | RUVA | | Authors: | Nishino, T, Ariyoshi, M, Iwasaki, H, Shinagawa, H, Morikawa, K. | | Deposit date: | 1997-08-21 | | Release date: | 1998-02-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Functional Analyses of the Domain Structure in the Holliday Junction Binding Protein Ruva
Structure, 6, 1998
|
|
1WYG
 
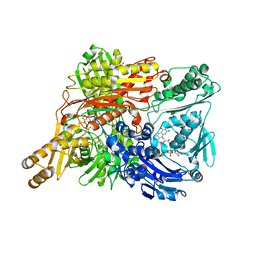 | | Crystal Structure of a Rat Xanthine Dehydrogenase Triple Mutant (C535A, C992R and C1324S) | | Descriptor: | 2-HYDROXYBENZOIC ACID, ACETIC ACID, CALCIUM ION, ... | | Authors: | Nishino, T, Okamoto, K, Kawaguchi, Y, Hori, H, Matsumura, T, Eger, B.T, Pai, E.F, Nishino, T. | | Deposit date: | 2005-02-14 | | Release date: | 2005-05-31 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanism of the Conversion of Xanthine Dehydrogenase to Xanthine Oxidase: IDENTIFICATION OF THE TWO CYSTEINE DISULFIDE BONDS AND CRYSTAL STRUCTURE OF A NON-CONVERTIBLE RAT LIVER XANTHINE DEHYDROGENASE MUTANT
J.Biol.Chem., 280, 2005
|
|
4YTZ
 
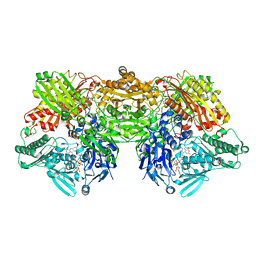 | | Rat xanthine oxidoreductase, C-terminal deletion protein variant, crystal grown without dithiothreitol | | Descriptor: | BICARBONATE ION, CALCIUM ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Nishino, T, Okamoto, K, Kawaguchi, Y, Matsumura, T, Eger, B.T, Pai, E.F, Nishino, T. | | Deposit date: | 2015-03-18 | | Release date: | 2015-04-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The C-terminal peptide plays a role in the formation of an intermediate form during the transition between xanthine dehydrogenase and xanthine oxidase
Febs J., 282, 2015
|
|
1IPI
 
 | |
1J23
 
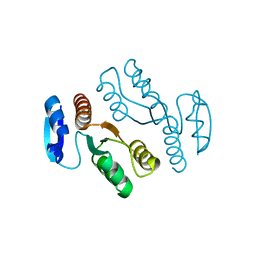 | | Crystal structure of archaeal XPF/Mus81 homolog, Hef from Pyrococcus furiosus, nuclease domain | | Descriptor: | ATP-dependent RNA helicase, putative | | Authors: | Nishino, T, Komori, K, Ishino, Y, Morikawa, K. | | Deposit date: | 2002-12-25 | | Release date: | 2003-04-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | X-Ray and Biochemical Anatomy of an Archaeal XPF/Rad1/Mus81 Family Nuclease. Similarity between Its Endonuclease Domain and Restriction Enzymes
Structure, 11, 2003
|
|
1J25
 
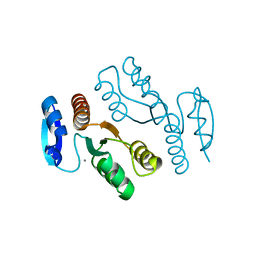 | | Crystal structure of archaeal XPF/Mus81 homolog, Hef from Pyrococcus furiosus, nuclease domain, Mn cocrystal | | Descriptor: | ATP-dependent RNA helicase, putative, MANGANESE (II) ION | | Authors: | Nishino, T, Komori, K, Ishino, Y, Morikawa, K. | | Deposit date: | 2002-12-25 | | Release date: | 2003-04-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | X-Ray and Biochemical Anatomy of an Archaeal XPF/Rad1/Mus81 Family Nuclease. Similarity between Its Endonuclease Domain and Restriction Enzymes
Structure, 11, 2003
|
|
1J22
 
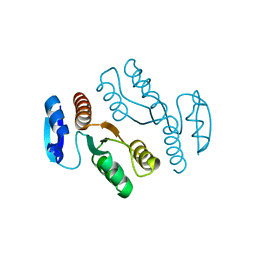 | | Crystal structure of archaeal XPF/Mus81 homolog, Hef from Pyrococcus furiosus, nuclease domain, selenomet derivative | | Descriptor: | ATP-dependent RNA helicase, putative | | Authors: | Nishino, T, Komori, K, Ishino, Y, Morikawa, K. | | Deposit date: | 2002-12-25 | | Release date: | 2003-04-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray and Biochemical Anatomy of an Archaeal XPF/Rad1/Mus81 Family Nuclease. Similarity between Its Endonuclease Domain and Restriction Enzymes
Structure, 11, 2003
|
|
1J24
 
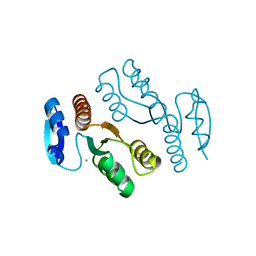 | | Crystal structure of archaeal XPF/Mus81 homolog, Hef from Pyrococcus furiosus, nuclease domain, Ca cocrystal | | Descriptor: | ATP-dependent RNA helicase, putative, CALCIUM ION | | Authors: | Nishino, T, Komori, K, Ishino, Y, Morikawa, K. | | Deposit date: | 2002-12-25 | | Release date: | 2003-04-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | X-Ray and Biochemical Anatomy of an Archaeal XPF/Rad1/Mus81 Family Nuclease. Similarity between Its Endonuclease Domain and Restriction Enzymes
Structure, 11, 2003
|
|
1D8L
 
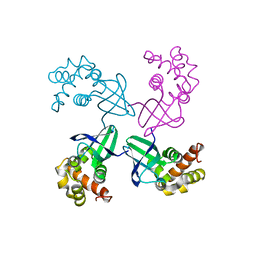 | | E. COLI HOLLIDAY JUNCTION BINDING PROTEIN RUVA NH2 REGION LACKING DOMAIN III | | Descriptor: | PROTEIN (HOLLIDAY JUNCTION DNA HELICASE RUVA) | | Authors: | Nishino, T, Iwasaki, H, Kataoka, M, Ariyoshi, M, Fujita, T, Shinagawa, H, Morikawa, K. | | Deposit date: | 1999-10-25 | | Release date: | 2000-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Modulation of RuvB function by the mobile domain III of the Holliday junction recognition protein RuvA.
J.Mol.Biol., 298, 2000
|
|
4YSW
 
 | | Structure of rat xanthine oxidoreductase, C-terminal deletion protein variant, NADH bound form | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, BICARBONATE ION, CALCIUM ION, ... | | Authors: | Nishino, T, Okamoto, K, Kawaguchi, Y, Matsumura, T, Eger, B.T, Pai, E.F. | | Deposit date: | 2015-03-17 | | Release date: | 2015-04-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The C-terminal peptide plays a role in the formation of an intermediate form during the transition between xanthine dehydrogenase and xanthine oxidase.
Febs J., 282, 2015
|
|
4YTY
 
 | | Structure of rat xanthine oxidoreductase, C535A/C992R/C1324S, NADH bound form | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, BICARBONATE ION, CALCIUM ION, ... | | Authors: | Nishino, T, Okamoto, K, Kawaguchi, Y, Matsumura, T, Eger, B.T, Pai, E.F. | | Deposit date: | 2015-03-18 | | Release date: | 2015-04-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The C-terminal peptide plays a role in the formation of an intermediate form during the transition between xanthine dehydrogenase and xanthine oxidase.
Febs J., 282, 2015
|
|
