5UGI
 
 | |
5TOO
 
 | | Crystal structure of alkaline phosphatase PafA T79S, N100A, K162A, R164A mutant | | Descriptor: | Alkaline phosphatase PafA, CHLORIDE ION, ZINC ION | | Authors: | Lyubimov, A.Y, Sunden, F, AlSadhan, I, Herschlag, D. | | Deposit date: | 2016-10-18 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.031 Å) | | Cite: | Differential catalytic promiscuity of the alkaline phosphatase superfamily bimetallo core reveals mechanistic features underlying enzyme evolution.
J. Biol. Chem., 292, 2017
|
|
6C17
 
 | | Crystal Structure of Ketosteroid Isomerase D40N mutant from Pseudomonas Putida (pKSI) bound to 3,4-dinitrophenol | | Descriptor: | 3,4-dinitrophenol, MAGNESIUM ION, Steroid Delta-isomerase | | Authors: | Yabukarski, F, Pinney, M, Herschlag, D. | | Deposit date: | 2018-01-04 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural Coupling Throughout the Active Site Hydrogen Bond Networks of Ketosteroid Isomerase and Photoactive Yellow Protein.
J. Am. Chem. Soc., 140, 2018
|
|
6C1J
 
 | | Crystal Structure of Ketosteroid Isomerase Y32F/Y57F/D40N mutant from Pseudomonas Putida (pKSI) bound to 3,4-dinitrophenol | | Descriptor: | 3,4-dinitrophenol, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Yabukarski, F, Pinney, M.M, Herschlag, D. | | Deposit date: | 2018-01-04 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.063 Å) | | Cite: | Structural Coupling Throughout the Active Site Hydrogen Bond Networks of Ketosteroid Isomerase and Photoactive Yellow Protein.
J. Am. Chem. Soc., 140, 2018
|
|
2GSO
 
 | | Structure of Xac Nucleotide Pyrophosphatase/Phosphodiesterase in Complex with Vanadate | | Descriptor: | VANADATE ION, ZINC ION, phosphodiesterase-nucleotide pyrophosphatase | | Authors: | Zalatan, J.G, Fenn, T.D, Brunger, A.T, Herschlag, D. | | Deposit date: | 2006-04-26 | | Release date: | 2006-08-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural and functional comparisons of nucleotide pyrophosphatase/phosphodiesterase and alkaline phosphatase: implications for mechanism and evolution
Biochemistry, 45, 2006
|
|
2GSU
 
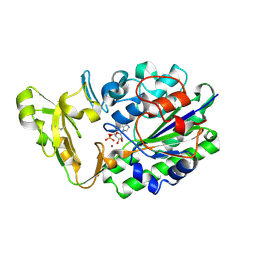 | | Structure of Xac Nucleotide Pyrophosphatase/Phosphodiesterase in Complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ZINC ION, phosphodiesterase-nucleotide pyrophosphatase | | Authors: | Zalatan, J.G, Fenn, T.D, Brunger, A.T, Herschlag, D. | | Deposit date: | 2006-04-26 | | Release date: | 2006-08-01 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional comparisons of nucleotide pyrophosphatase/phosphodiesterase and alkaline phosphatase: implications for mechanism and evolution
Biochemistry, 45, 2006
|
|
2GSN
 
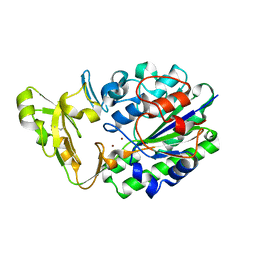 | | Structure of Xac Nucleotide Pyrophosphatase/Phosphodiesterase | | Descriptor: | ZINC ION, phosphodiesterase-nucleotide pyrophosphatase | | Authors: | Zalatan, J.G, Fenn, T.D, Brunger, A.T, Herschlag, D. | | Deposit date: | 2006-04-26 | | Release date: | 2006-08-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and functional comparisons of nucleotide pyrophosphatase/phosphodiesterase and alkaline phosphatase: implications for mechanism and evolution
Biochemistry, 45, 2006
|
|
3OWS
 
 | |
8F01
 
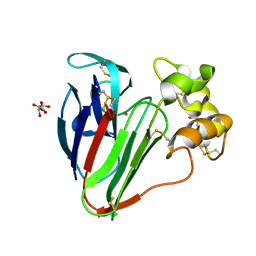 | |
8F05
 
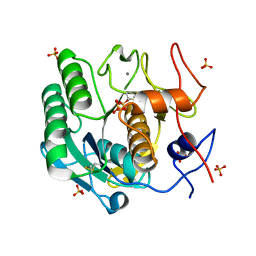 | |
8EZX
 
 | | Lysozyme Anomalous Dataset at 293 K and 7.1 keV | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Doukov, T, Yabukarski, F, Herschlag, D. | | Deposit date: | 2022-11-01 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Obtaining anomalous and ensemble information from protein crystals from 220 K up to physiological temperatures.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8F06
 
 | |
8F07
 
 | |
8F0B
 
 | | Lysozyme Anomalous Dataset at 240 K and 7.1 keV | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Doukov, T, Yabukarski, F, Herschlag, D. | | Deposit date: | 2022-11-02 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Obtaining anomalous and ensemble information from protein crystals from 220 K up to physiological temperatures.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8F03
 
 | | Thaumatin Anomalous Dataset at 293 K and 12 keV | | Descriptor: | L(+)-TARTARIC ACID, POTASSIUM ION, Thaumatin I | | Authors: | Doukov, T, Yabukarski, F, Herschlag, D. | | Deposit date: | 2022-11-01 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Obtaining anomalous and ensemble information from protein crystals from 220 K up to physiological temperatures.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8F00
 
 | | Lysozyme Anomalous Dataset at 293 K and 12 keV | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Doukov, T, Yabukarski, F, Herschlag, D. | | Deposit date: | 2022-11-01 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Obtaining anomalous and ensemble information from protein crystals from 220 K up to physiological temperatures.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8EZP
 
 | | Lysozyme Anomalous Dataset at 260 K and 7.1 keV | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Doukov, T, Yabukarski, F, Herschlag, D. | | Deposit date: | 2022-11-01 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Obtaining anomalous and ensemble information from protein crystals from 220 K up to physiological temperatures.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8EZU
 
 | | Lysozyme Anomalous Dataset at 273 K and 7.1 keV | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Doukov, T, Yabukarski, F, Herschlag, D. | | Deposit date: | 2022-11-01 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Obtaining anomalous and ensemble information from protein crystals from 220 K up to physiological temperatures.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8SOV
 
 | | Proteinase K Multiconformer Model at 353K | | Descriptor: | ALA-ALA-ALA-SER-VAL-LYS, CALCIUM ION, Proteinase K, ... | | Authors: | Du, S, Wankowicz, S, Yabukarski, F, Doukov, T, Herschlag, D, Fraser, J.S. | | Deposit date: | 2023-04-30 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.291 Å) | | Cite: | Refinement of multiconformer ensemble models from multi-temperature X-ray diffraction data.
Methods Enzymol., 688, 2023
|
|
8SPL
 
 | | Proteinase K Multiconformer Model at 343K | | Descriptor: | ALA-ALA-ALA-SER-VAL-LYS, CALCIUM ION, Proteinase K, ... | | Authors: | Du, S, Wankowicz, S, Yabukarski, F, Doukov, T, Herschlag, D, Fraser, J.S. | | Deposit date: | 2023-05-03 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Refinement of multiconformer ensemble models from multi-temperature X-ray diffraction data.
Methods Enzymol., 688, 2023
|
|
8SOG
 
 | | Proteinase K Multiconformer Model at 313K | | Descriptor: | CALCIUM ION, Proteinase K, SULFATE ION | | Authors: | Du, S, Wankowicz, S, Yabukarski, F, Doukov, T, Herschlag, D, Fraser, J.S. | | Deposit date: | 2023-04-28 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Refinement of multiconformer ensemble models from multi-temperature X-ray diffraction data.
Methods Enzymol., 688, 2023
|
|
8SOU
 
 | | Proteinase K Multiconformer Model at 363K | | Descriptor: | ALA-ALA-ALA-SER-VAL-LYS, CALCIUM ION, Proteinase K, ... | | Authors: | Du, S, Wankowicz, S, Yabukarski, F, Doukov, T, Herschlag, D, Fraser, J.S. | | Deposit date: | 2023-04-30 | | Release date: | 2023-08-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Refinement of multiconformer ensemble models from multi-temperature X-ray diffraction data.
Methods Enzymol., 688, 2023
|
|
8SQV
 
 | | Proteinase K Multiconformer Model at 333K | | Descriptor: | CALCIUM ION, Proteinase K, SULFATE ION | | Authors: | Du, S, Wankowicz, S, Yabukarski, F, Doukov, T, Herschlag, D, Fraser, J.S. | | Deposit date: | 2023-05-04 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Refinement of multiconformer ensemble models from multi-temperature X-ray diffraction data.
Methods Enzymol., 688, 2023
|
|
3TG0
 
 | | E. coli alkaline phosphatase with bound inorganic phosphate | | Descriptor: | Alkaline phosphatase, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Bobyr, E, Lassila, J.K, Wiersma-Koch, H.I, Fenn, T.D, Lee, J.J, Nikolic-Hughes, I, Hodgson, K.O, Rees, D.C, Hedman, B, Herschlag, D. | | Deposit date: | 2011-08-16 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High-resolution analysis of Zn(2+) coordination in the alkaline phosphatase superfamily by EXAFS and x-ray crystallography.
J.Mol.Biol., 415, 2012
|
|
3T8N
 
 | | Crystal structure of ketosteroid isomerase Y16AD103A from Pseudomonas putida | | Descriptor: | SULFATE ION, Steroid Delta-isomerase, {[-(BIS-CARBOXYMETHYL-AMINO)-ETHYL]-CARBOXYMETHYL-AMINO}-ACETIC ACID | | Authors: | Gonzalez, A, Tsai, Y, Schwans, J, Sunden, F, Herschlag, D. | | Deposit date: | 2011-08-01 | | Release date: | 2011-11-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Evaluating the catalytic contribution from the oxyanion hole in ketosteroid isomerase.
J.Am.Chem.Soc., 133, 2011
|
|
