5T3K
 
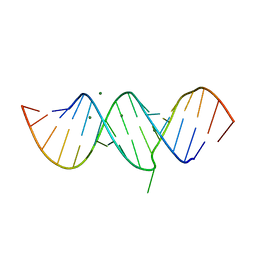 | |
2OE6
 
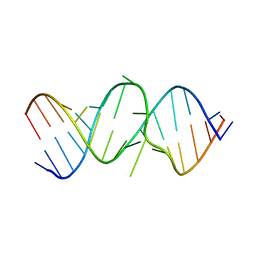 | | 2.4A X-ray crystal structure of unliganded RNA fragment GGGCGUCGCUAGUACC/CGGUACUAAAAGUCGCC containing the human ribosomal decoding A site: RNA construct with 5'-overhang | | Descriptor: | RNA (5'-R(*CP*GP*GP*UP*AP*CP*UP*AP*AP*AP*AP*GP*UP*CP*GP*CP*C)-3'), RNA (5'-R(*GP*GP*GP*CP*GP*UP*CP*GP*CP*UP*AP*GP*UP*AP*CP*C)-3') | | Authors: | Hermann, T, Tereshko, V, Skripkin, E, Patel, D.J. | | Deposit date: | 2006-12-28 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Apramycin recognition by the human ribosomal decoding site.
Blood Cells Mol.Dis., 38, 2007
|
|
2OE8
 
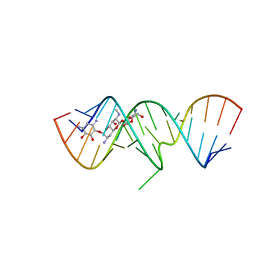 | | 1.8 A X-ray crystal structure of Apramycin complex with RNA fragment GGGCGUCGCUAGUACC/CGGUACUAAAAGUCGCC containing the human ribosomal decoding A site: RNA construct with 5'-overhang | | Descriptor: | APRAMYCIN, RNA (5'-R(*CP*GP*GP*UP*AP*CP*UP*AP*AP*AP*AP*GP*UP*CP*GP*CP*C)-3'), RNA (5'-R(*GP*GP*GP*CP*GP*UP*CP*GP*CP*UP*AP*GP*UP*AP*CP*C)-3') | | Authors: | Hermann, T, Tereshko, V, Skripkin, E, Patel, D.J. | | Deposit date: | 2006-12-28 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Apramycin recognition by the human ribosomal decoding site.
Blood Cells Mol.Dis., 38, 2007
|
|
2OE5
 
 | | 1.5 A X-ray crystal structure of Apramycin complex with RNA fragment GGCGUCGCUAGUACCG/GGUACUAAAAGUCGCCC containing the human ribosomal decoding A site: RNA construct with 3'-overhang | | Descriptor: | APRAMYCIN, MAGNESIUM ION, RNA (5'-R(*GP*GP*CP*GP*UP*CP*GP*CP*UP*AP*GP*UP*AP*CP*CP*G)-3'), ... | | Authors: | Hermann, T, Tereshko, V, Skripkin, E, Patel, D.J. | | Deposit date: | 2006-12-28 | | Release date: | 2007-02-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Apramycin recognition by the human ribosomal decoding site.
Blood Cells Mol.Dis., 38, 2007
|
|
1EEG
 
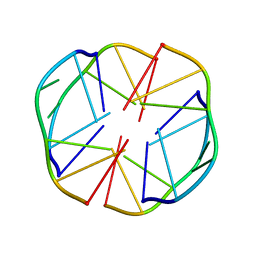 | | A(GGGG)A HEXAD PAIRING ALIGMENT FOR THE D(G-G-A-G-G-A-G) SEQUENCE | | Descriptor: | DNA (5'-D(*GP*GP*AP*GP*GP*A)-3') | | Authors: | Kettani, A, Gorin, A, Majumdar, A, Hermann, T, Skripkin, E, Zhao, H, Jones, R, Patel, D.J. | | Deposit date: | 2000-01-31 | | Release date: | 2000-04-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A dimeric DNA interface stabilized by stacked A.(G.G.G.G).A hexads and coordinated monovalent cations.
J.Mol.Biol., 297, 2000
|
|
3P59
 
 | | First Crystal Structure of a RNA Nanosquare | | Descriptor: | COBALT HEXAMMINE(III), MAGNESIUM ION, RNA (5'-R(*CP*CP*GP*GP*AP*GP*GP*AP*AP*CP*UP*AP*CP*(5BU)P*G)-3'), ... | | Authors: | Dibrov, S, Hermann, T, McLean, J, Parsons, J. | | Deposit date: | 2010-10-08 | | Release date: | 2011-04-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1793 Å) | | Cite: | Self-assembling RNA square.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
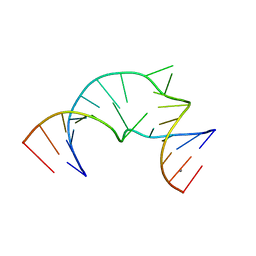
4P97
 
 | |
4PHY
 
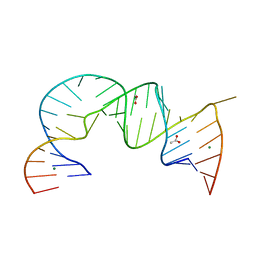 | | Functional conservation despite structural divergence in ligand-responsive RNA switches | | Descriptor: | ACETATE ION, MAGNESIUM ION, RNA (26-MER), ... | | Authors: | Boerneke, M.A, Dibrov, S.M, Hermann, T.H. | | Deposit date: | 2014-05-07 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Functional conservation despite structural divergence in ligand-responsive RNA switches.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3MEI
 
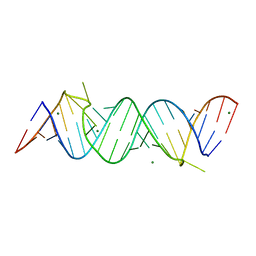 | | Regulatory motif from the thymidylate synthase mRNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*CP*CP*GP*CP*CP*GP*CP*GP*CP*CP*AP*(5BU)P*GP*CP*CP*UP*GP*UP*GP*GP*CP*GP*G)-3') | | Authors: | Dibrov, S, McLean, J, Hermann, T. | | Deposit date: | 2010-03-31 | | Release date: | 2011-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.968 Å) | | Cite: | Structure of an RNA dimer of a regulatory element from human thymidylate synthase mRNA.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
5CNR
 
 | |
1G70
 
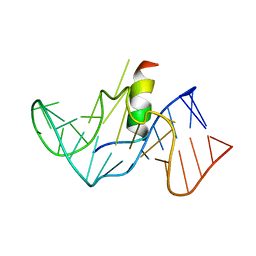 | | COMPLEX OF HIV-1 RRE-IIB RNA WITH RSG-1.2 PEPTIDE | | Descriptor: | HIV-1 RRE-IIB 32 NUCLEOTIDE RNA, RSG-1.2 PEPTIDE | | Authors: | Gosser, Y, Hermann, T, Majumdar, A, Hu, W, Frederick, R, Jiang, F, Xu, W, Patel, D.J. | | Deposit date: | 2000-11-08 | | Release date: | 2001-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Peptide-triggered conformational switch in HIV-1 RRE RNA complexes.
Nat.Struct.Biol., 8, 2001
|
|
3TZR
 
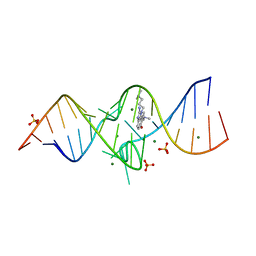 | | Structure of a Riboswitch-like RNA-ligand complex from the Hepatitis C Virus Internal Ribosome Entry Site | | Descriptor: | (8R)-8-[(dimethylamino)methyl]-1-[3-(dimethylamino)propyl]-1,7,8,9-tetrahydrochromeno[5,6-d]imidazol-2-amine, 5'-R(*CP*GP*AP*GP*GP*AP*AP*CP*UP*AP*CP*UP*GP*UP*CP*UP*UP*CP*CP*C)-3', 5'-R(*GP*GP*UP*CP*GP*UP*GP*CP*AP*GP*CP*CP*UP*CP*GP*G)-3', ... | | Authors: | Dibrov, S.M, Ding, K, Brunn, N, Parker, M.A, Bergdahl, B.M, Wyles, D.L, Hermann, T. | | Deposit date: | 2011-09-27 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.212 Å) | | Cite: | Structure of a Riboswitch in the Hepatitis C Virus Internal Ribosome Entry Site
To be Published
|
|
2PN3
 
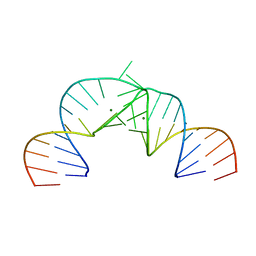 | | Crystal Structure of Hepatitis C Virus IRES Subdomain IIa | | Descriptor: | 5'-R(*CP*GP*GP*AP*GP*GP*AP*AP*CP*UP*AP*CP*UP*GP*UP*CP*UP*UP*CP*AP*CP*GP*CP*C)-3', 5'-R(*GP*CP*GP*(5BU)P*GP*UP*CP*GP*UP*GP*CP*AP*GP*CP*CP*(5BU)P*CP*CP*GP*G)-3', MAGNESIUM ION | | Authors: | Zhao, Q, Han, Q, Kissinger, C.R, Hermann, T, Thompson, P.A. | | Deposit date: | 2007-04-23 | | Release date: | 2008-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of hepatitis C virus IRES subdomain IIa.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
1T0D
 
 | | Crystal Structure of 2-aminopurine labelled bacterial decoding site RNA | | Descriptor: | 5'-R(*CP*AP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*AP*CP*CP*C)-3', 5'-R(*GP*GP*UP*GP*GP*UP*GP*(MTU)P*AP*GP*UP*CP*GP*CP*UP*GP*G)-3' | | Authors: | Shandrick, S, Zhao, Q, Han, Q, Ayida, B.K, Takahashi, M, Winters, G.C, Simonsen, K.B, Vourloumis, D, Hermann, T. | | Deposit date: | 2004-04-08 | | Release date: | 2004-06-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Monitoring molecular recognition of the ribosomal decoding site.
Angew.Chem.Int.Ed.Engl., 43, 2004
|
|
1T0E
 
 | | Crystal Structure of 2-aminopurine labelled bacterial decoding site RNA | | Descriptor: | 5'-R(*CP*GP*AP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*AP*CP*CP*C)-3', 5'-R(*GP*GP*UP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*CP*UP*CP*GP*G)-3', SULFATE ION | | Authors: | Shandrick, S, Zhao, Q, Han, Q, Ayida, B.K, Takahashi, M, Winters, G.C, Simonsen, K.B, Vourloumis, D, Hermann, T. | | Deposit date: | 2004-04-08 | | Release date: | 2004-06-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Monitoring molecular recognition of the ribosomal decoding site.
Angew.Chem.Int.Ed.Engl., 43, 2004
|
|
4GYH
 
 | |
4H1I
 
 | |
1YRJ
 
 | | Crystal Structure of Apramycin bound to a Ribosomal RNA A site oligonucleotide | | Descriptor: | APRAMYCIN, Bacterial 16 S Ribosomal RNA A Site Oligonucleotide, MAGNESIUM ION, ... | | Authors: | Han, Q, Zhao, Q, Fish, S, Simonsen, K.B, Vourloumis, D, Froelich, J.M, Wall, D, Hermann, T. | | Deposit date: | 2005-02-03 | | Release date: | 2005-06-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular recognition by glycoside pseudo base pairs and triples in an apramycin-RNA complex.
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
2A04
 
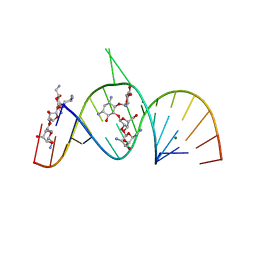 | | Molecular Recognition of RNA by Neomycin and a Restricted Neomycin Derivative | | Descriptor: | 5'-R(*CP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*AP*CP*C)-3', 5'-R(*GP*UP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*CP*GP*G)-3', MAGNESIUM ION, ... | | Authors: | Zhao, Q, Zhao, F, Blount, K.F, Han, Q, Tor, Y, Hermann, T. | | Deposit date: | 2005-06-15 | | Release date: | 2005-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Molecular recognition of RNA by neomycin and a restricted neomycin derivative
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
1ZX7
 
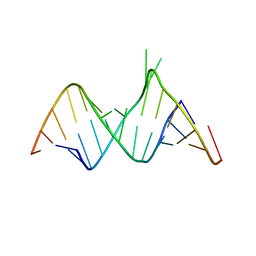 | | Molecular Recognition of RNA by Neomycin and a Restricted Neomycin Derivative | | Descriptor: | 5'-R(*CP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*AP*CP*C)-3', 5'-R(*GP*UP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*CP*GP*G)-3' | | Authors: | Zhao, Q, Zhao, F, Blount, K.F, Han, Q, Tor, Y, Hermann, T. | | Deposit date: | 2005-06-07 | | Release date: | 2005-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular recognition of RNA by neomycin and a restricted neomycin derivative
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
1ZZ5
 
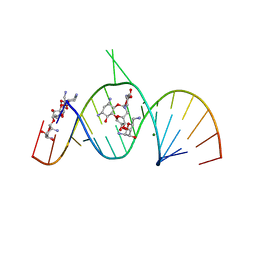 | | Molecular Recognition of RNA by Neomycin and a Restricted Neomycin Derivative | | Descriptor: | 13,15-DIAMINO-2-(AMINOMETHYL)-3,4,9,12-TETRAHYDROXYHEXADECAHYDRO-2H-7,10-EPOXYPYRANO[2,3-B][1,10,4]BENZODIOXAZACYCLODODECIN-8-YL 2,6-DIAMINO-2,6-DIDEOXYHEXOPYRANOSIDE, 5'-R(*CP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*AP*CP*C)-3', 5'-R(*GP*UP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*CP*GP*G)-3', ... | | Authors: | Zhao, Q, Zhao, F, Blount, K.F, Han, Q, Tor, Y, Hermann, T. | | Deposit date: | 2005-06-13 | | Release date: | 2005-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular recognition of RNA by neomycin and a restricted neomycin derivative
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
8ADD
 
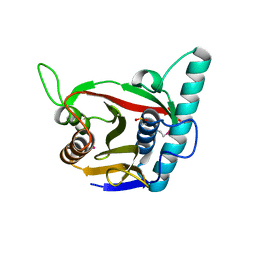 | | Viral tegument-like DUBs | | Descriptor: | ATP-dependent DNA helicase | | Authors: | Erven, I, Abraham, E.T, Hermanns, T, Hofmann, K, Baumann, U. | | Deposit date: | 2022-07-08 | | Release date: | 2023-02-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A widely distributed family of eukaryotic and bacterial deubiquitinases related to herpesviral large tegument proteins.
Nat Commun, 13, 2022
|
|
8CMR
 
 | | Linear specific OTU-type DUB SnOTU from the pathogen S. negenvensis in complex with linear di-ubiquitin | | Descriptor: | OTU domain-containing protein, Polyubiquitin-B | | Authors: | Uthoff, M, Hermanns, T, Boll, V, Hofmann, K, Baumann, U. | | Deposit date: | 2023-02-21 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Functional and structural diversity in deubiquitinases of the Chlamydia-like bacterium Simkania negevensis.
Nat Commun, 14, 2023
|
|
7OJE
 
 | | Crystal structure of the covalent complex between Tribolium castaneum deubiquitinase ZUP and Ubiquitin-PA | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, Lys-63-specific deubiquitinase ZUFSP, ... | | Authors: | Pichlo, C, Hermanns, T, Hofmann, K, Baumann, U. | | Deposit date: | 2021-05-14 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A structural basis for the diverse linkage specificities within the ZUFSP deubiquitinase family.
Nat Commun, 13, 2022
|
|
7OIY
 
 | | Crystal structure of the ZUFSP family member Mug105 | | Descriptor: | SODIUM ION, Ubiquitin carboxyl-terminal hydrolase mug105 | | Authors: | Pichlo, C, Hermanns, T, Hofmann, K, Baumann, U. | | Deposit date: | 2021-05-12 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A structural basis for the diverse linkage specificities within the ZUFSP deubiquitinase family.
Nat Commun, 13, 2022
|
|
