2VB7
 
 | | beta-ketoacyl-ACP synthase I (KAS) from E. coli, apo structure after soak in PEG solution | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] SYNTHASE 1 | | Authors: | Pappenberger, G, Schulz-Gasch, T, Bailly, J, Hennig, M. | | Deposit date: | 2007-09-06 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Assisted Discovery of an Aminothiazole Derivative as a Lead Molecule for Inhibition of Bacterial Fatty-Acid Synthesis.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2VBA
 
 | | beta-ketoacyl-ACP synthase I (KAS) from E. coli with bound amino- thiazole inhibitor | | Descriptor: | 2-PHENYLAMINO-4-METHYL-5-ACETYL THIAZOLE, 3-OXOACYL-[ACYL-CARRIER-PROTEIN] SYNTHASE 1 | | Authors: | Pappenberger, G, Schulz-Gasch, T, Bailly, J, Hennig, M. | | Deposit date: | 2007-09-06 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structure-Assisted Discovery of an Aminothiazole Derivative as a Lead Molecule for Inhibition of Bacterial Fatty-Acid Synthesis.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2VB8
 
 | | beta-ketoacyl-ACP synthase I (KAS) from E. coli with bound inhibitor thiolactomycin | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] SYNTHASE 1, CHLORIDE ION, THIOLACTOMYCIN | | Authors: | Pappenberger, G, Schulz-Gasch, T, Bailly, J, Hennig, M. | | Deposit date: | 2007-09-06 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structure-Assisted Discovery of an Aminothiazole Derivative as a Lead Molecule for Inhibition of Bacterial Fatty-Acid Synthesis.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
6DM7
 
 | |
6DXM
 
 | |
6DVT
 
 | |
6ED9
 
 | |
4AFZ
 
 | | Human Chymase - Fynomer Complex | | Descriptor: | CHYMASE, D(-)-TARTARIC ACID, FYNOMER | | Authors: | Schlatter, D, Brack, S, Banner, D.W, Batey, S, Benz, J, Bertschinger, J, Huber, W, Joseph, C, Rufer, A, Van Der Kloosters, A, Weber, M, Grabulovski, D, Hennig, M. | | Deposit date: | 2012-01-23 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Generation, Characterization and Structural Data of Chymase Binding Proteins Based on the Human Fyn Kinase SH3 Domain.
Mabs, 4, 2012
|
|
4AFQ
 
 | | Human Chymase - Fynomer Complex | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CHYMASE, CITRATE ANION, ... | | Authors: | Schlatter, D, Brack, S, Banner, D.W, Batey, S, Benz, J, Bertschinger, J, Huber, W, Joseph, C, Rufer, A, Van Der Kloosters, A, Weber, M, Grabulovski, D, Hennig, M. | | Deposit date: | 2012-01-23 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Generation, Characterization and Structural Data of Chymase Binding Proteins Based on the Human Fyn Kinase SH3 Domain.
Mabs, 4, 2012
|
|
4AFS
 
 | | Human Chymase - Fynomer Complex | | Descriptor: | CHYMASE, FYNOMER, GLYCEROL, ... | | Authors: | Schlatter, D, Brack, S, Banner, D.W, Batey, S, Benz, J, Bertschinger, J, Huber, W, Joseph, C, Rufer, A, Van Der Kloosters, A, Weber, M, Grabulovski, D, Hennig, M. | | Deposit date: | 2012-01-23 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Generation, Characterization and Structural Data of Chymase Binding Proteins Based on the Human Fyn Kinase SH3 Domain.
Mabs, 4, 2012
|
|
4AG2
 
 | | Human Chymase - Fynomer Complex | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHYMASE, ... | | Authors: | Schlatter, D, Brack, S, Banner, D.W, Batey, S, Benz, J, Bertschinger, J, Huber, W, Joseph, C, Rufer, A, Van Der Kloosters, A, Weber, M, Grabulovski, D, Hennig, M. | | Deposit date: | 2012-01-23 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Generation, Characterization and Structural Data of Chymase Binding Proteins Based on the Human Fyn Kinase SH3 Domain.
Mabs, 4, 2012
|
|
4AG1
 
 | | Human Chymase - Fynomer Complex | | Descriptor: | CHYMASE, FYNOMER, SULFATE ION | | Authors: | Schlatter, D, Brack, S, Banner, D.W, Batey, S, Benz, J, Bertschinger, J, Huber, W, Joseph, C, Rufer, A, Van Der Kloosters, A, Weber, M, Grabulovski, D, Hennig, M. | | Deposit date: | 2012-01-23 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Generation, Characterization and Structural Data of Chymase Binding Proteins Based on the Human Fyn Kinase SH3 Domain.
Mabs, 4, 2012
|
|
4AFU
 
 | | Human Chymase - Fynomer Complex | | Descriptor: | CHYMASE, FYNOMER | | Authors: | Schlatter, D, Brack, S, Banner, D.W, Batey, S, Benz, J, Bertschinger, J, Huber, W, Joseph, C, Rufer, A, Van Der Kloosters, A, Weber, M, Grabulovski, D, Hennig, M. | | Deposit date: | 2012-01-23 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Generation, Characterization and Structural Data of Chymase Binding Proteins Based on the Human Fyn Kinase SH3 Domain.
Mabs, 4, 2012
|
|
8CIC
 
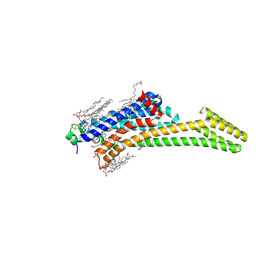 | | Crystal structure of stabilized A2A adenosine receptor A2AR-StaR2-bRIL in complex with clinical candidate Etrumadenant | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-[2-azanyl-6-[1-[[6-(2-oxidanylpropan-2-yl)pyridin-2-yl]methyl]-1,2,3-triazol-4-yl]pyrimidin-4-yl]-2-methyl-benzenecarbonitrile, ... | | Authors: | Cheng, R.K.Y, Markovic-Mueller, S, Hennig, M. | | Deposit date: | 2023-02-09 | | Release date: | 2023-05-31 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Crystal structure of adenosine A 2A receptor in complex with clinical candidate Etrumadenant reveals unprecedented antagonist interaction.
Commun Chem, 6, 2023
|
|
2RP0
 
 | |
