3O1J
 
 | |
3O1I
 
 | |
7RAX
 
 | |
3PRA
 
 | |
3PRD
 
 | |
3OFE
 
 | |
8FM0
 
 | | HIV-1 gp120 complex with CJF-III-214 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, methyl (2R,3S)-2-(carbamimidamidomethyl)-3-[2-(4-chloro-3-fluoroanilino)(oxo)acetamido]-6-[(methylamino)methyl]-2,3-dihydro-1H-indole-1-carboxylate | | Authors: | Gong, Z, Hendrickson, W.A. | | Deposit date: | 2022-12-22 | | Release date: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Indoline CD4-mimetic compounds mediate potent and broad HIV-1 inhibition and sensitization to antibody-dependent cellular cytotoxicity.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FM4
 
 | | HIV-1 gp120 complex with CJF-IV-047 | | Descriptor: | 2,2,2-trifluoroethyl (2R,3S)-2-(carbamimidamidomethyl)-3-[2-(4-chloro-3-fluoroanilino)(oxo)acetamido]-6-[(methylamino)methyl]-2,3-dihydro-1H-indole-1-carboxylate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120 | | Authors: | Gong, Z, Hendrickson, W.A. | | Deposit date: | 2022-12-22 | | Release date: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Indoline CD4-mimetic compounds mediate potent and broad HIV-1 inhibition and sensitization to antibody-dependent cellular cytotoxicity.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FLZ
 
 | | HIV-1 gp120 complex with CJF-III-049-S | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, N~1~-{(1R,2R,3S)-2-(carbamimidamidomethyl)-3-[(3S)-3,4-dihydroxybutyl]-5-[(methylamino)methyl]-2,3-dihydro-1H-inden-1-yl}-N~2~-(4-chloro-3-fluorophenyl)ethanediamide | | Authors: | Gong, Z, Hendrickson, W.A. | | Deposit date: | 2022-12-22 | | Release date: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Indoline CD4-mimetic compounds mediate potent and broad HIV-1 inhibition and sensitization to antibody-dependent cellular cytotoxicity.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FM5
 
 | | HIV-1 gp120 complex with DY-III-065 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,3,3-trifluoropropyl (2R,3S)-2-(carbamimidamidomethyl)-3-[2-(4-chloro-3-fluoroanilino)(oxo)acetamido]-6-[(methylamino)methyl]-2,3-dihydro-1H-indole-1-carboxylate, Envelope glycoprotein gp120 | | Authors: | Gong, Z, Hendrickson, W.A. | | Deposit date: | 2022-12-22 | | Release date: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Indoline CD4-mimetic compounds mediate potent and broad HIV-1 inhibition and sensitization to antibody-dependent cellular cytotoxicity.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FM7
 
 | | HIV-1 gp120 complex with CJF-III-192 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, benzyl (2R,3S)-2-(carbamimidamidomethyl)-3-[2-(4-chloro-3-fluoroanilino)(oxo)acetamido]-6-[(methylamino)methyl]-2,3-dihydro-1H-indole-1-carboxylate | | Authors: | Gong, Z, Hendrickson, W.A. | | Deposit date: | 2022-12-22 | | Release date: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Indoline CD4-mimetic compounds mediate potent and broad HIV-1 inhibition and sensitization to antibody-dependent cellular cytotoxicity.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FLY
 
 | | HIV-1 gp120 complex with BNM-III-170 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ~{N}'-[(1~{R},2~{R})-2-(carbamimidamidomethyl)-5-(methylaminomethyl)-2,3-dihydro-1~{H}-inden-1-yl]-~{N}-(4-chloranyl-3-fluoranyl-phenyl)ethanediamide | | Authors: | Gong, Z, Hendrickson, W.A. | | Deposit date: | 2022-12-22 | | Release date: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Indoline CD4-mimetic compounds mediate potent and broad HIV-1 inhibition and sensitization to antibody-dependent cellular cytotoxicity.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FM2
 
 | | HIV-1 gp120 complex with CJF-III-289 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, IMIDAZOLE, ... | | Authors: | Gong, Z, Hendrickson, W.A. | | Deposit date: | 2022-12-22 | | Release date: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Indoline CD4-mimetic compounds mediate potent and broad HIV-1 inhibition and sensitization to antibody-dependent cellular cytotoxicity.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FM3
 
 | | HIV-1 gp120 complex with CJF-III-288 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, propyl (2R,3S)-2-(carbamimidamidomethyl)-3-[2-(4-chloro-3-fluoroanilino)(oxo)acetamido]-6-[(methylamino)methyl]-2,3-dihydro-1H-indole-1-carboxylate | | Authors: | Gong, Z, Hendrickson, W.A. | | Deposit date: | 2022-12-22 | | Release date: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Indoline CD4-mimetic compounds mediate potent and broad HIV-1 inhibition and sensitization to antibody-dependent cellular cytotoxicity.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FM8
 
 | | HIV-1 gp120 complex with CJF-IV-046 | | Descriptor: | (pentafluorophenyl)methyl (2R,3S)-2-(carbamimidamidomethyl)-3-[2-(4-chloro-3-fluoroanilino)(oxo)acetamido]-6-[(methylamino)methyl]-2,3-dihydro-1H-indole-1-carboxylate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Gong, Z, Hendrickson, W.A. | | Deposit date: | 2022-12-22 | | Release date: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Indoline CD4-mimetic compounds mediate potent and broad HIV-1 inhibition and sensitization to antibody-dependent cellular cytotoxicity.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3OFH
 
 | | Structured Domain of Mus musculus Mesd | | Descriptor: | LDLR chaperone MESD | | Authors: | Collins, M.N, Hendrickson, W.A. | | Deposit date: | 2010-08-14 | | Release date: | 2011-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.013 Å) | | Cite: | Structural Characterization of the Boca/Mesd Maturation Factors for LDL-Receptor-Type beta-Propeller Domains
Structure, 2011
|
|
3PR9
 
 | |
3PRB
 
 | |
3OFF
 
 | |
3OFG
 
 | | Structured Domain of Caenorhabditis elegans BMY-1 | | Descriptor: | Boca/mesd chaperone for ywtd beta-propeller-egf protein 1, CHLORIDE ION | | Authors: | Collins, M.N, Hendrickson, W.A. | | Deposit date: | 2010-08-14 | | Release date: | 2011-03-02 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.367 Å) | | Cite: | Structural Characterization of the Boca/Mesd Maturation Factors for LDL-Receptor-Type beta-Propeller Domains
Structure, 2011
|
|
3OTT
 
 | |
1CDU
 
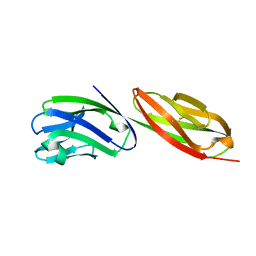 | | STRUCTURE OF T-CELL SURFACE GLYCOPROTEIN CD4 MUTANT WITH PHE 43 REPLACED BY VAL | | Descriptor: | T-CELL SURFACE GLYCOPROTEIN CD4 | | Authors: | Wu, H, Myszka, D, Tendian, S.W, Brouillette, C.G, Sweet, R.W, Chaiken, I.M, Hendrickson, W.A. | | Deposit date: | 1996-11-11 | | Release date: | 1997-04-01 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Kinetic and structural analysis of mutant CD4 receptors that are defective in HIV gp120 binding.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1CDJ
 
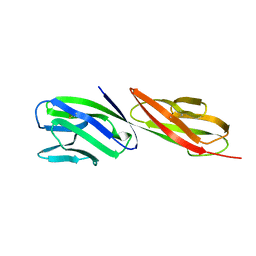 | | STRUCTURE OF T-CELL SURFACE GLYCOPROTEIN CD4 | | Descriptor: | T-CELL SURFACE GLYCOPROTEIN CD4 | | Authors: | Wu, H, Myszka, D, Tendian, S.W, Brouillette, C.G, Sweet, R.W, Chaiken, I.M, Hendrickson, W.A. | | Deposit date: | 1996-11-11 | | Release date: | 1997-04-01 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Kinetic and structural analysis of mutant CD4 receptors that are defective in HIV gp120 binding.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1CDY
 
 | | STRUCTURE OF T-CELL SURFACE GLYCOPROTEIN CD4 MUTANT WITH GLY 47 REPLACED BY SER | | Descriptor: | T-CELL SURFACE GLYCOPROTEIN CD4 | | Authors: | Wu, H, Myszka, D, Tendian, S.W, Brouillette, C.G, Sweet, R.W, Chaiken, I.M, Hendrickson, W.A. | | Deposit date: | 1996-11-11 | | Release date: | 1997-04-01 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and structural analysis of mutant CD4 receptors that are defective in HIV gp120 binding.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1RZJ
 
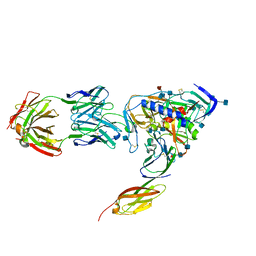 | | HIV-1 HXBC2 GP120 ENVELOPE GLYCOPROTEIN COMPLEXED WITH CD4 AND INDUCED NEUTRALIZING ANTIBODY 17B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY 17B, HEAVY CHAIN, ... | | Authors: | Huang, C.C, Venturi, M, Majeed, S, Moore, M.J, Phogat, S, Zhang, M.-Y, Dimitrov, D.S, Hendrickson, W.A, Robinson, J, Sodroski, J, Wyatt, R, Choe, H, Farzan, M, Kwong, P.D. | | Deposit date: | 2003-12-24 | | Release date: | 2004-02-03 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
