3BTW
 
 | | THE CRYSTAL STRUCTURES OF THE COMPLEXES BETWEEN BOVINE BETA-TRYPSIN AND TEN P1 VARIANTS OF BPTI | | Descriptor: | CALCIUM ION, PROTEIN (BOVINE PANCREATIC TRYPSIN INHIBITOR), PROTEIN (TRYPSIN), ... | | Authors: | Helland, R, Otlewski, J, Sundheim, O, Dadlez, M, Smalas, A.O. | | Deposit date: | 1999-03-11 | | Release date: | 2000-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The crystal structures of the complexes between bovine beta-trypsin and ten P1 variants of BPTI.
J.Mol.Biol., 287, 1999
|
|
3BTF
 
 | | THE CRYSTAL STRUCTURES OF THE COMPLEXES BETWEEN BOVINE BETA-TRYPSIN AND TEN P1 VARIANTS OF BPTI. | | Descriptor: | CALCIUM ION, PROTEIN (PANCREATIC TRYPSIN INHIBITOR), PROTEIN (TRYPSIN), ... | | Authors: | Helland, R, Otlewski, J, Sundheim, O, Dadlez, M, Smalas, A.O. | | Deposit date: | 1999-03-10 | | Release date: | 2000-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structures of the complexes between bovine beta-trypsin and ten P1 variants of BPTI.
J.Mol.Biol., 287, 1999
|
|
3BTM
 
 | | THE CRYSTAL STRUCTURES OF THE COMPLEXES BETWEEN BOVINE BETA-TRYPSIN AND TEN P1 VARIANTS OF BPTI | | Descriptor: | CALCIUM ION, PROTEIN (PANCREATIC TRYPSIN INHIBITOR), PROTEIN (TRYPSIN), ... | | Authors: | Helland, R, Otlewski, J, Sundheim, O, Dadlez, M, Smalas, A.O. | | Deposit date: | 1999-03-10 | | Release date: | 2000-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structures of the complexes between bovine beta-trypsin and ten P1 variants of BPTI.
J.Mol.Biol., 287, 1999
|
|
3BTD
 
 | | The Crystal Structures of the Complexes Between the Bovine Beta-Trypsin and Ten P1 Variants of BPTI. | | Descriptor: | CALCIUM ION, PROTEIN (BOVINE PANCREATIC TRYPSIN INHIBITOR), PROTEIN (TRYPSIN), ... | | Authors: | Helland, R, Otlewski, J, Sundheim, O, Dadlez, M, Smalas, A.O. | | Deposit date: | 1999-03-11 | | Release date: | 2000-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structures of the complexes between bovine beta-trypsin and ten P1 variants of BPTI.
J.Mol.Biol., 287, 1999
|
|
3BTK
 
 | | THE CRYSTAL STRUCTURES OF THE COMPLEXES BETWEEN BOVINE BETA-TRYPSIN AND TEN P1 VARIANTS OF BPTI | | Descriptor: | CALCIUM ION, PROTEIN (PANCREATIC TRYPSIN INHIBITOR), PROTEIN (TRYPSIN), ... | | Authors: | Helland, R, Otlewski, J, Sundheim, O, Dadlez, M, Smalas, A.O. | | Deposit date: | 1999-03-10 | | Release date: | 2000-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structures of the complexes between bovine beta-trypsin and ten P1 variants of BPTI.
J.Mol.Biol., 287, 1999
|
|
3BTE
 
 | | The Crystal Structures of the Complexes Between Bovine Beta-Trypsin and Ten P1 Variants of BPTI. | | Descriptor: | CALCIUM ION, PANCREATIC TRYPSIN INHIBITOR, SULFATE ION, ... | | Authors: | Helland, R, Otlewski, J, Sundheim, O, Dadlez, M, Smalas, A.O. | | Deposit date: | 1999-03-11 | | Release date: | 2000-03-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structures of the complexes between bovine beta-trypsin and ten P1 variants of BPTI.
J.Mol.Biol., 287, 1999
|
|
3BTG
 
 | | THE CRYSTAL STRUCTURES OF THE COMPLEXES BETWEEN BOVINE BETA-TRYPSIN AND TEN P1 VARIANTS OF BPTI | | Descriptor: | CALCIUM ION, PROTEIN (PANCREATIC TRYPSIN INHIBITOR), PROTEIN (TRYPSIN), ... | | Authors: | Helland, R, Otlewski, J, Sundheim, O, Dadlez, M, Smalas, A.O. | | Deposit date: | 1999-03-10 | | Release date: | 2000-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structures of the complexes between bovine beta-trypsin and ten P1 variants of BPTI.
J.Mol.Biol., 287, 1999
|
|
1EJM
 
 | | CRYSTAL STRUCTURE OF THE BPTI ALA16LEU MUTANT IN COMPLEX WITH BOVINE TRYPSIN | | Descriptor: | BETA-TRYPSIN, PANCREATIC TRYPSIN INHIBITOR, SULFATE ION | | Authors: | Otlewski, J, Smalas, A, Helland, R, Grzesiak, A, Krowarsch, D. | | Deposit date: | 2000-03-03 | | Release date: | 2001-03-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substitutions at the P(1) position in BPTI strongly affect the association energy with serine proteinases.
J.Mol.Biol., 301, 2000
|
|
6XYZ
 
 | | Crystal structure of the GH18 chitinase ChiB from the chitin utilization locus of Flavobacterium johnsoniae | | Descriptor: | 1,2-ETHANEDIOL, Candidate chitinase Glycoside hydrolase family 18, FORMIC ACID | | Authors: | Mazurkewich, S, Helland, R, MacKenzie, A, Eijsink, V, Pope, P, Branden, G, Larsbrink, J. | | Deposit date: | 2020-01-31 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural insights of the enzymes from the chitin utilization locus of Flavobacterium johnsoniae.
Sci Rep, 10, 2020
|
|
5ACX
 
 | | VIM-2-2, Discovery of novel inhibitor scaffolds against the metallo- beta-lactamase VIM-2 by SPR based fragment screening | | Descriptor: | 2-(4-fluorophenyl)carbonylbenzoic acid, BETA-LACTAMASE, CHLORIDE ION, ... | | Authors: | Christopeit, T, Carlsen, T.J.O, Helland, R, Leiros, H.K.S. | | Deposit date: | 2015-08-18 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Novel Inhibitor Scaffolds Against the Metallo-Beta-Lactamase Vim-2 by Spr Based Fragment Screening
J.Med.Chem., 58, 2015
|
|
5ACW
 
 | | VIM-2-1, Discovery of novel inhibitor scaffolds against the metallo- beta-lactamase VIM-2 by SPR based fragment screening | | Descriptor: | 4-methyl-5-(trifluoromethyl)-1,2,4-triazole-3-thiol, BETA-LACTAMASE, CHLORIDE ION, ... | | Authors: | Christopeit, T, Carlsen, T.J.O, Helland, R, Leiros, H.K.S. | | Deposit date: | 2015-08-18 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Novel Inhibitor Scaffolds Against the Metallo-Beta-Lactamase Vim-2 by Spr Based Fragment Screening
J.Med.Chem., 58, 2015
|
|
2G7F
 
 | | The 1.95 A crystal structure of Vibrio cholerae extracellular endonuclease I | | Descriptor: | CHLORIDE ION, Endonuclease I, MAGNESIUM ION | | Authors: | Altermark, B, Smalaas, A.O, Willassen, N.P, Helland, R. | | Deposit date: | 2006-02-28 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structure of Vibrio cholerae extracellular endonuclease I reveals the presence of a buried chloride ion.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2G7E
 
 | | The 1.6 A crystal structure of Vibrio cholerae extracellular endonuclease I | | Descriptor: | CHLORIDE ION, Endonuclease I | | Authors: | Altermark, B, Smalaas, A.O, Willassen, N.P, Helland, R. | | Deposit date: | 2006-02-28 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of Vibrio cholerae extracellular endonuclease I reveals the presence of a buried chloride ion.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
6QSQ
 
 | |
6YHH
 
 | | X-ray Structure of Flavobacterium johnsoniae chitobiase (FjGH20) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-N-acetylglucosaminidase-like protein Glycoside hydrolase family 20, GLYCEROL | | Authors: | Mazurkewich, S, Helland, R, MacKenzie, A, Eijsink, V.G.H, Pope, P.B, Branden, G, Larsbrink, J. | | Deposit date: | 2020-03-30 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights of the enzymes from the chitin utilization locus of Flavobacterium johnsoniae.
Sci Rep, 10, 2020
|
|
6T26
 
 | | X-ray crystal structure of Vibrio alkaline phosphatase with the non-competitive inhibitor cyclohexylamine | | Descriptor: | Alkaline phosphatase, CHLORIDE ION, CYCLOHEXYLAMMONIUM ION, ... | | Authors: | Asgeirsson, B, Hjorleifsson, J.G, Markusson, S, Helland, R. | | Deposit date: | 2019-10-07 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.265 Å) | | Cite: | X-ray crystal structure of Vibrio alkaline phosphatase with the non-competitive inhibitor cyclohexylamine.
Biochem Biophys Rep, 24, 2020
|
|
4BZ4
 
 | | CorA is a surface-associated copper-binding protein important in Methylomicrobium album BG8 copper acquisition | | Descriptor: | CALCIUM ION, COPPER (I) ION, COPPER-REPRESSIBLE POLYPEPTIDE, ... | | Authors: | Johnson, K.A, Ve, T, Pedersen, R.B, Lillehaug, J.R, Jensen, H.B, Helland, R, Karlsen, O.A. | | Deposit date: | 2013-07-24 | | Release date: | 2014-02-19 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cora is a Copper Repressible Surface-Associated Copper(I)-Binding Protein Produced in Methylomicrobium Album Bg8.
Plos One, 9, 2014
|
|
3MGW
 
 | | Thermodynamics and structure of a salmon cold-active goose-type lysozyme | | Descriptor: | COBALT (II) ION, Lysozyme g, SULFATE ION | | Authors: | Kyomuhendo, P, Myrnes, B, Brandsdal, B.O, Smalas, A.O, Nilsen, I.W, Helland, R. | | Deposit date: | 2010-04-07 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Thermodynamics and structure of a salmon cold active goose-type lysozyme
Comp.Biochem.Physiol. B: Biochem.Mol.Biol., 156, 2010
|
|
2ISA
 
 | | Crystal Structure of Vibrio salmonicida catalase | | Descriptor: | CHLORIDE ION, Catalase, GLYCEROL, ... | | Authors: | Riise, E.K, Lorentzen, M.S, Helland, R, Smalas, A.O, Leiros, H.K.S, Willassen, N.P. | | Deposit date: | 2006-10-17 | | Release date: | 2007-01-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The first structure of a cold-active catalase from Vibrio salmonicida at 1.96A reveals structural aspects of cold adaptation
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
5AFD
 
 | | Native structure of N-acetylneuramininate lyase (sialic acid aldolase) from Aliivibrio salmonicida | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, N-ACETYLNEURAMINATE LYASE | | Authors: | Gurung, M.K, Altermark, B, Rader, I.L.U, Helland, R, Smalas, A.O. | | Deposit date: | 2015-01-21 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Features and structure of a cold active N-acetylneuraminate lyase.
Plos One, 14, 2019
|
|
5ACU
 
 | | VIM-2-NAT, Discovery of novel inhibitor scaffolds against the metallo- beta-lactamase VIM-2 by SPR based fragment screening | | Descriptor: | BETA-LACTAMASE, CHLORIDE ION, HYDROXIDE ION, ... | | Authors: | Christopeit, T, Carlsen, T.J.O, Helland, R, Leiros, H.K.S. | | Deposit date: | 2015-08-18 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of Novel Inhibitor Scaffolds Against the Metallo-Beta-Lactamase Vim-2 by Spr Based Fragment Screening
J.Med.Chem., 58, 2015
|
|
5ACV
 
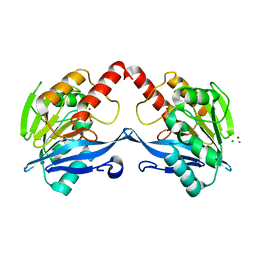 | | VIM-2-OX, Discovery of novel inhibitor scaffolds against the metallo- beta-lactamase VIM-2 by SPR based fragment screening | | Descriptor: | BETA-LACTAMASE, CHLORIDE ION, HYDROXIDE ION, ... | | Authors: | Christopeit, T, Carlsen, T.J.O, Helland, R, Leiros, H.K.S. | | Deposit date: | 2015-08-18 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.963 Å) | | Cite: | Discovery of Novel Inhibitor Scaffolds Against the Metallo-Beta-Lactamase Vim-2 by Spr Based Fragment Screening
J.Med.Chem., 58, 2015
|
|
2PU3
 
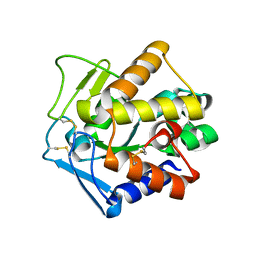 | | Structural adaptation of endonuclease I from the cold-adapted and halophilic bacterium Vibrio salmonicida | | Descriptor: | CHLORIDE ION, Endonuclease I, MAGNESIUM ION | | Authors: | Altermark, B, Helland, R, Moe, E, Willassen, N.P, Smalas, A.O. | | Deposit date: | 2007-05-08 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural adaptation of endonuclease I from the cold-adapted and halophilic bacterium Vibrio salmonicida.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2Q9X
 
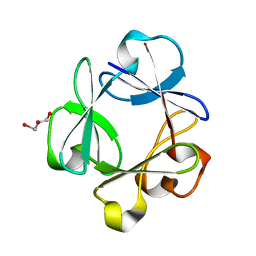 | | Crystal structure of highly stable mutant Q40P/S47I/H93G of human fibroblast growth factor-1 | | Descriptor: | GLYCEROL, Heparin-binding growth factor 1 | | Authors: | Szlachcic, A, Zakrzewska, M, Krowarsch, D, Os, V, Helland, R, Otlewski, J. | | Deposit date: | 2007-06-14 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of highly stable mutant Q40P/S47I/H93G of human fibroblast growth factor-1
To be Published
|
|
4OY8
 
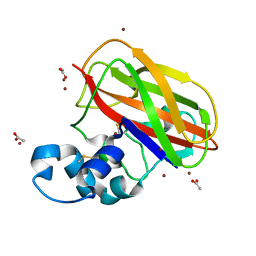 | | Structure of ScLPMO10B in complex with zinc. | | Descriptor: | ACETATE ION, Putative secreted cellulose-binding protein, ZINC ION | | Authors: | Forsberg, Z, Mackenzie, A.K, Sorlie, M, Rohr, A.K, Helland, R, Arvai, A.S, Vaaje-Kolstad, G, Eijsink, V.G.H. | | Deposit date: | 2014-02-11 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and functional characterization of a conserved pair of bacterial cellulose-oxidizing lytic polysaccharide monooxygenases.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
