3SZS
 
 | |
1W7R
 
 | | Feglymycin P64 crystal form | | Descriptor: | CITRATE ANION, FEGLYMYCIN, ISOPROPYL ALCOHOL | | Authors: | Bunkoczi, G, Vertesy, L, Sheldrick, G.M. | | Deposit date: | 2004-09-08 | | Release date: | 2005-03-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Antiviral Antibiotic Feglymycin: First Direct Methods Solution of a 1000Plus Equal-Atom Structure
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
2QC7
 
 | | Crystal structure of the protein-disulfide isomerase related chaperone ERp29 | | Descriptor: | Endoplasmic reticulum protein ERp29 | | Authors: | Barak, N.N, Sevvana, M, Neumann, P, Malesevic, M, Naumann, K, Fischer, G, Sheldrick, G.M, Stubbs, M.T, Ferrari, D.M. | | Deposit date: | 2007-06-19 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure and functional analysis of the protein disulfide isomerase-related protein ERp29.
J.Mol.Biol., 385, 2009
|
|
1LZ8
 
 | | LYSOZYME PHASED ON ANOMALOUS SIGNAL OF SULFURS AND CHLORINES | | Descriptor: | CHLORIDE ION, PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Dauter, Z, Dauter, M, De La Fortelle, E, Bricogne, G, Sheldrick, G.M. | | Deposit date: | 1999-03-14 | | Release date: | 1999-05-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Can anomalous signal of sulfur become a tool for solving protein crystal structures?
J.Mol.Biol., 289, 1999
|
|
1BX7
 
 | | HIRUSTASIN FROM HIRUDO MEDICINALIS AT 1.2 ANGSTROMS | | Descriptor: | HIRUSTASIN, SULFATE ION | | Authors: | Uson, I, Sheldrick, G.M, De La Fortelle, E, Bricogne, G, Di Marco, S, Priestle, J.P, Gruetter, M.G, Mittl, P.R.E. | | Deposit date: | 1998-10-14 | | Release date: | 1999-04-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The 1.2 A crystal structure of hirustasin reveals the intrinsic flexibility of a family of highly disulphide-bridged inhibitors.
Structure Fold.Des., 7, 1999
|
|
4MIV
 
 | | Crystal Structure of Sulfamidase, Crystal Form L | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Sidhu, N.S, Uson, I, Schreiber, K, Proepper, K, Becker, S, Sheldrick, G.M, Gaertner, J, Kraetzner, R, Steinfeld, R. | | Deposit date: | 2013-09-02 | | Release date: | 2014-05-14 | | Last modified: | 2021-06-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of sulfamidase provides insight into the molecular pathology of mucopolysaccharidosis IIIA.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1W3M
 
 | | Crystal structure of tsushimycin | | Descriptor: | CALCIUM ION, CHLORIDE ION, Delta-3isotetradecenoic acid, ... | | Authors: | Bunkoczi, G, Vertesy, L, Sheldrick, G.M. | | Deposit date: | 2004-07-16 | | Release date: | 2005-07-27 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure of the lipopeptide antibiotic tsushimycin.
Acta Crystallogr. D Biol. Crystallogr., 61, 2005
|
|
3SIL
 
 | | SIALIDASE FROM SALMONELLA TYPHIMURIUM | | Descriptor: | GLYCEROL, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Garman, E.F, Sheldrick, G.M. | | Deposit date: | 1998-07-07 | | Release date: | 1998-11-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | An Enzyme at Atomic Resolution: The 1.05 A Structure of Salmonella Typhimurium Neuraminidase (Sialidase)
To be Published
|
|
1BX8
 
 | | HIRUSTASIN FROM HIRUDO MEDICINALIS AT 1.4 ANGSTROMS | | Descriptor: | HIRUSTASIN, SULFATE ION | | Authors: | Uson, I, Sheldrick, G.M, De La Fortelle, E, Bricogne, G, Di Marco, S, Priestle, J.P, Gruetter, M.G, Mittl, P.R.E. | | Deposit date: | 1998-10-14 | | Release date: | 1999-04-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The 1.2 A crystal structure of hirustasin reveals the intrinsic flexibility of a family of highly disulphide-bridged inhibitors.
Structure Fold.Des., 7, 1999
|
|
1UNM
 
 | | Crystal structure of 7-Aminoactinomycin D with non-complementary DNA | | Descriptor: | 5'-D(*TP*TP*AP*GP*BRU*TP)-3', 7-AMINOACTINOMYCIN D | | Authors: | Alexopoulos, E.C, Klement, R, Jares-Erijman, E.A, Uson, I, Jovin, T.M, Sheldrick, G.M. | | Deposit date: | 2003-09-11 | | Release date: | 2004-09-24 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal and Solution Structures of 7-Amino-Actinomycin D Complexes with D(Ttagbrut), D(Ttagtt) and D(Tttagttt)
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1W7Q
 
 | | Feglymycin P65 crystal form | | Descriptor: | 1,2-ETHANEDIOL, ETHANOL, FEGLYMYCIN | | Authors: | Bunkoczi, G, Vertesy, L, Sheldrick, G.M. | | Deposit date: | 2004-09-08 | | Release date: | 2005-03-08 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The Antiviral Antibiotic Feglymycin: First Direct Methods Solution of a 1000Plus Equal-Atom Structure
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
1VTR
 
 | | STRUCTURE OF THE DEOXYTETRANUCLEOTIDE D-PAPTPAPT AND A SEQUENCE-DEPENDENT MODEL FOR POLY(DA-DT) | | Descriptor: | DNA (5'-D(*AP*TP*AP*T)-3') | | Authors: | Viswamitra, M.A, Shakked, Z, Jones, P.G, Sheldrick, G.M, Salisbury, S.A, Kennard, O. | | Deposit date: | 1988-08-18 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Structure of the Deoxytetranucleotide d-pApTpApT and a Sequence-Dependent Model for Poly(dA-dT)
Biopolymers, 21, 1982
|
|
3ITI
 
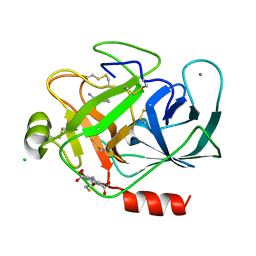 | | Structure of bovine trypsin with the MAD triangle B3C | | Descriptor: | 5-amino-2,4,6-tribromobenzene-1,3-dicarboxylic acid, BENZAMIDINE, CALCIUM ION, ... | | Authors: | Beck, T, da Cunha, C.E, Sheldrick, G.M. | | Deposit date: | 2009-08-28 | | Release date: | 2009-10-27 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | How to get the magic triangle and the MAD triangle into your protein crystal.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
6OR0
 
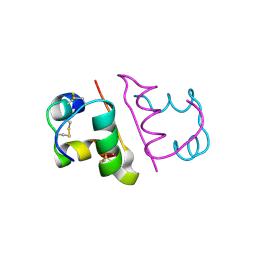 | | Crystal structure of Insulin from Non-merohedrally twinned crystals | | Descriptor: | Insulin chain A, Insulin chain B | | Authors: | Sevvana, M, Ruf, M, Uson, I, Sheldrick, G.M, Herbst-Irmer, R. | | Deposit date: | 2019-04-29 | | Release date: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Non-merohedral twinning: from minerals to proteins.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
3JU4
 
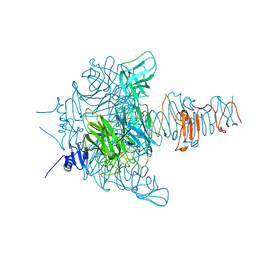 | | Crystal Structure Analysis of EndosialidaseNF at 0.98 A Resolution | | Descriptor: | CHLORIDE ION, Endo-N-acetylneuraminidase, N-acetyl-beta-neuraminic acid, ... | | Authors: | Schulz, E.C, Neuman, P, Gerardy-Schahn, R, Sheldrick, G.M, Ficner, R. | | Deposit date: | 2009-09-14 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Structure analysis of endosialidase NF at 0.98 A resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
6OQZ
 
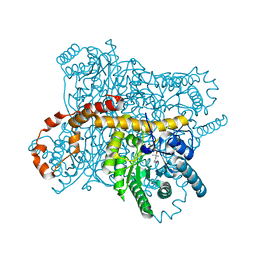 | | Crystal structure of Glucose Isomerase from Non-merohedrally twinned crystals | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Sevvana, M, Ruf, M, Uson, I, Sheldrick, G.M, Herbst-Irmer, R. | | Deposit date: | 2019-04-29 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Non-merohedral twinning: from minerals to proteins.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
1HH3
 
 | | Decaplanin first P21-Form | | Descriptor: | 4-epi-vancosamine, DECAPLANIN, GLYCEROL, ... | | Authors: | Lehmann, C, Vertessy, L, Sheldrick, G.M, Dauter, Z, Dauter, M. | | Deposit date: | 2000-12-19 | | Release date: | 2005-07-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structures of Four Crystal Forms of Decaplanin
Helv.Chim.Acta, 86, 2003
|
|
1HHF
 
 | | Decaplanin second P6122-Form | | Descriptor: | 4-epi-vancosamine, CHLORIDE ION, DECAPLANIN, ... | | Authors: | Lehmann, C, Vertessy, L, Sheldrick, G.M, Dauter, Z, Dauter, M. | | Deposit date: | 2000-12-22 | | Release date: | 2005-07-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structures of Four Crystal Forms of Decaplanin
Helv.Chim.Acta, 86, 2003
|
|
1HHA
 
 | | Decaplanin first P6122-Form | | Descriptor: | 4-epi-vancosamine, DECAPLANIN, GLYCEROL, ... | | Authors: | Lehmann, C, Vertessy, L, Sheldrick, G.M, Dauter, Z, Dauter, M. | | Deposit date: | 2000-12-22 | | Release date: | 2005-07-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Four Crystal Forms of Decaplanin
Helv.Chim.Acta, 86, 2003
|
|
1H34
 
 | | Crystal structure of lima bean trypsin inhibitor | | Descriptor: | BOWMAN-BIRK TYPE PROTEINASE INHIBITOR | | Authors: | Debreczeni, J.E, Bunkoczi, G, Girmann, B, Sheldrick, G.M. | | Deposit date: | 2002-08-21 | | Release date: | 2003-02-06 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | In-House Phase Determination of the Lima Bean Trypsin Inhibitor: A Low-Resolution Sulfur-Sad Case
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1HHC
 
 | | Crystal structure of Decaplanin - space group P21, second form | | Descriptor: | 4-epi-vancosamine, CITRIC ACID, DECAPLANIN, ... | | Authors: | Lehmann, C, Vertessy, L, Sheldrick, G.M, Dauter, Z, Dauter, M. | | Deposit date: | 2000-12-22 | | Release date: | 2005-07-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Structures of Four Crystal Forms of Decaplanin
Helv.Chim.Acta, 86, 2003
|
|
1LU0
 
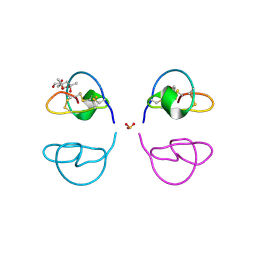 | | Atomic Resolution Structure of Squash Trypsin Inhibitor: Unexpected Metal Coordination | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Thaimattam, R, Tykarska, E, Bierzynski, A, Sheldrick, G.M, Jaskolski, M. | | Deposit date: | 2002-05-21 | | Release date: | 2002-08-28 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Atomic resolution structure of squash trypsin inhibitor: unexpected metal coordination.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
4JRD
 
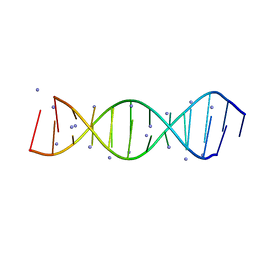 | | Crystal structure of the parallel double-stranded helix of poly(A) RNA | | Descriptor: | AMMONIUM ION, RNA (5'-R(*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3') | | Authors: | Safaee, N, Noronha, A.M, Kozlov, G, Rodionov, D, Wilds, C.J, Sheldrick, G.M, Gehring, K. | | Deposit date: | 2013-03-21 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure of the parallel duplex of poly(A) RNA: evaluation of a 50 year-old prediction.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
2ADW
 
 | |
1OB6
 
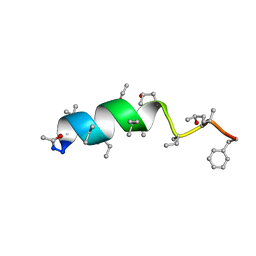 | | Cephaibol B | | Descriptor: | ACETATE ION, CEPHAIBOL B, ETHANOL | | Authors: | Bunkoczi, G, Schiell, M, Vertesy, L, Sheldrick, G.M. | | Deposit date: | 2003-01-24 | | Release date: | 2003-12-11 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Crystal Structures of Cephaibols
J.Pept.Sci., 9, 2003
|
|
