1O6V
 
 | | Internalin (INLA, Listeria monocytogenes) - functional domain, uncomplexed | | Descriptor: | CALCIUM ION, INTERNALIN A | | Authors: | Schubert, W.-D, Urbanke, C, Ziehm, T, Beier, V, Machner, M.P, Domann, E, Wehland, J, Chakraborty, T, Heinz, D.W. | | Deposit date: | 2002-10-16 | | Release date: | 2002-12-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Internalin, a Major Invasion Protein of Listeria Monocytogenes, in Complex with its Human Receptor E-Cadherin
Cell(Cambridge,Mass.), 111, 2002
|
|
2OMV
 
 | | Crystal structure of InlA S192N Y369S/hEC1 complex | | Descriptor: | CALCIUM ION, CHLORIDE ION, Epithelial-cadherin, ... | | Authors: | Wollert, T, Heinz, D.W, Schubert, W.D. | | Deposit date: | 2007-01-23 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Extending the host range of Listeria monocytogenes by rational protein design.
Cell(Cambridge,Mass.), 129, 2007
|
|
1OLT
 
 | | Coproporphyrinogen III oxidase (HemN) from Escherichia coli is a Radical SAM enzyme. | | Descriptor: | IRON/SULFUR CLUSTER, OXYGEN-INDEPENDENT COPROPORPHYRINOGEN III OXIDASE, S-ADENOSYLMETHIONINE | | Authors: | Layer, G, Moser, J, Heinz, D.W, Jahn, D, Schubert, W.-D. | | Deposit date: | 2003-08-13 | | Release date: | 2003-12-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal Structure of Coproporphyrinogen III Oxidase Reveals Cofactor Geometry of Radical Sam Enzymes
Embo J., 22, 2003
|
|
2OMY
 
 | | Crystal structure of InlA S192N/hEC1 complex | | Descriptor: | CALCIUM ION, CHLORIDE ION, Epithelial-cadherin, ... | | Authors: | Wollert, T, Heinz, D.W, Schubert, W.D. | | Deposit date: | 2007-01-23 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Extending the host range of Listeria monocytogenes by rational protein design.
Cell(Cambridge,Mass.), 129, 2007
|
|
2OMW
 
 | | Crystal structure of InlA S192N Y369S/mEC1 complex | | Descriptor: | CHLORIDE ION, Epithelial-cadherin, Internalin-A | | Authors: | Wollert, T, Heinz, D.W, Schubert, W.D. | | Deposit date: | 2007-01-23 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Extending the host range of Listeria monocytogenes by rational protein design.
Cell(Cambridge,Mass.), 129, 2007
|
|
2OMX
 
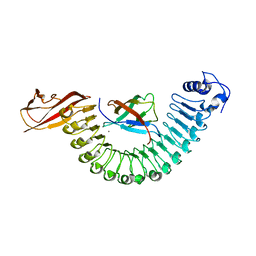 | | Crystal structure of InlA S192N G194S+S/hEC1 complex | | Descriptor: | CALCIUM ION, CHLORIDE ION, Epithelial-cadherin, ... | | Authors: | Wollert, T, Heinz, D.W, Schubert, W.D. | | Deposit date: | 2007-01-23 | | Release date: | 2007-08-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Thermodynamically reengineering the listerial invasion complex InlA/E-cadherin.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2OMZ
 
 | |
2OMT
 
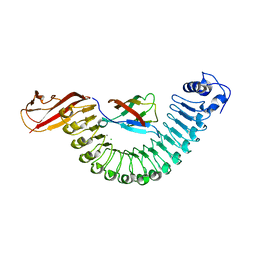 | | Crystal structure of InlA G194S+S/hEC1 complex | | Descriptor: | CALCIUM ION, CHLORIDE ION, Epithelial-cadherin; E-Cad/CTF1, ... | | Authors: | Wollert, T, Heinz, D.W, Schubert, W.D. | | Deposit date: | 2007-01-23 | | Release date: | 2007-08-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Thermodynamically reengineering the listerial invasion complex InlA/E-cadherin.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4A1T
 
 | | Co-Complex of the of NS3-4A protease with the inhibitory peptide CP5- 46-A (in-House data) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BICARBONATE ION, CHLORIDE ION, ... | | Authors: | Schmelz, S, Kuegler, J, Collins, J, Heinz, D.W. | | Deposit date: | 2011-09-19 | | Release date: | 2012-09-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | High Affinity Peptide Inhibitors of the Hepatitis C Virus Ns3-4A Protease Refractory to Common Resistant Mutants.
J.Biol.Chem., 287, 2012
|
|
4A10
 
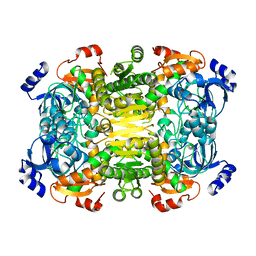 | | Apo-structure of 2-octenoyl-CoA carboxylase reductase CinF from streptomyces sp. | | Descriptor: | OCTENOYL-COA REDUCTASE/CARBOXYLASE | | Authors: | Quade, N, Huo, L, Rachid, S, Heinz, D.W, Muller, R. | | Deposit date: | 2011-09-13 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Unusual Carbon Fixation Giving Rise to Diverse Polyketide Extender Units
Nat.Chem.Biol., 8, 2011
|
|
4A1V
 
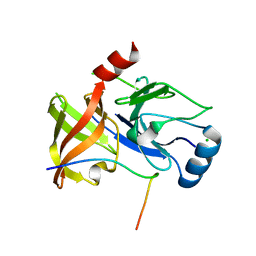 | | Co-Complex structure of NS3-4A protease with the optimized inhibitory peptide CP5-46A-4D5E | | Descriptor: | CHLORIDE ION, CP5-46A-4D5E, NON-STRUCTURAL PROTEIN 4A, ... | | Authors: | Schmelz, S, Kuegler, J, Collins, J, Heinz, D.W. | | Deposit date: | 2011-09-20 | | Release date: | 2012-09-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High Affinity Peptide Inhibitors of the Hepatitis C Virus Ns3-4A Protease Refractory to Common Resistant Mutants.
J.Biol.Chem., 287, 2012
|
|
4AW4
 
 | |
2OMU
 
 | | Crystal structure of InlA G194S+S Y369S/hEC1 complex | | Descriptor: | CALCIUM ION, CHLORIDE ION, Epithelial-cadherin, ... | | Authors: | Wollert, T, Heinz, D.W, Schubert, W.D. | | Deposit date: | 2007-01-23 | | Release date: | 2007-08-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Thermodynamically reengineering the listerial invasion complex InlA/E-cadherin.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1SBN
 
 | |
2UZX
 
 | | Structure of the human receptor tyrosine kinase Met in complex with the Listeria monocytogenes invasion protein InlB: Crystal form I | | Descriptor: | HEPATOCYTE GROWTH FACTOR RECEPTOR, INTERNALIN B | | Authors: | Niemann, H.H, Jager, V, Butler, P.J.G, Van Den Heuvel, J, Schmidt, S, Ferraris, D, Gherardi, E, Heinz, D.W. | | Deposit date: | 2007-05-02 | | Release date: | 2007-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Human Receptor Tyrosine Kinase met in Complex with the Listeria Invasion Protein Inlb
Cell(Cambridge,Mass.), 130, 2007
|
|
1O6T
 
 | | Internalin (INLA, Listeria monocytogenes) - functional domain, uncomplexed | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Schubert, W.-D, Urbanke, C, Ziehm, T, Beier, V, Machner, M.P, Domann, E, Wehland, J, Chakraborty, T, Heinz, D.W. | | Deposit date: | 2002-10-15 | | Release date: | 2002-12-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Internalin, a Major Invasion Protein of Listeria Monocytogenes, in Complex with its Human Receptor E-Cadherin
Cell(Cambridge,Mass.), 111, 2002
|
|
1SIB
 
 | |
2UZY
 
 | | Structure of the human receptor tyrosine kinase Met in complex with the Listeria monocytogenes invasion protein inlb: low resolution, Crystal form II | | Descriptor: | HEPATOCYTE GROWTH FACTOR RECEPTOR, INTERNALIN B | | Authors: | Niemann, H.H, Jager, V, Butler, P.J.G, van den Heuvel, J, Schmidt, S, Ferraris, D, Gherardi, E, Heinz, D.W. | | Deposit date: | 2007-05-02 | | Release date: | 2007-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structure of the Human Receptor Tyrosine Kinase met in Complex with the Listeria Invasion Protein Inlb
Cell(Cambridge,Mass.), 130, 2007
|
|
2V3A
 
 | | Crystal structure of rubredoxin reductase from Pseudomonas aeruginosa. | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Hagelueken, G, Wiehlmann, L, Adams, T.M, Kolmar, H, Heinz, D.W, Tuemmler, B, Schubert, W.-D. | | Deposit date: | 2007-06-14 | | Release date: | 2007-08-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Electron Transfer Complex Rubredoxin - Rubredoxin Reductase from Pseudomonas Aeruginosa.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2V3B
 
 | | Crystal structure of the electron transfer complex rubredoxin - rubredoxin reductase from Pseudomonas aeruginosa. | | Descriptor: | FE (III) ION, FLAVIN-ADENINE DINUCLEOTIDE, RUBREDOXIN 2, ... | | Authors: | Hagelueken, G, Wiehlmann, L, Adams, T.M, Kolmar, H, Heinz, D.W, Tuemmler, B, Schubert, W.-D. | | Deposit date: | 2007-06-14 | | Release date: | 2007-08-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of the Electron Transfer Complex Rubredoxin - Rubredoxin Reductase from Pseudomonas Aeruginosa.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
4CH7
 
 | | Crystal structure of the siroheme decarboxylase NirDL | | Descriptor: | NIRD-LIKE PROTEIN | | Authors: | Schmelz, S, Kriegler, T.M, Haufschildt, K, Layer, G, Heinz, D.W. | | Deposit date: | 2013-11-29 | | Release date: | 2014-07-30 | | Last modified: | 2014-09-10 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | The Crystal Structure of Siroheme Decarboxylase in Complex with Iron-Uroporphyrin III Reveals Two Essential Histidine Residues
J.Mol.Biol., 426, 2014
|
|
2VGX
 
 | | Structure of the Yersinia enterocolitica Type III Secretion Translocator Chaperone SycD | | Descriptor: | CHAPERONE SYCD | | Authors: | Buttner, C.R, Sorg, I, Cornelis, G.R, Heinz, D.W, Niemann, H.H. | | Deposit date: | 2007-11-16 | | Release date: | 2007-12-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the Yersinia Enterocolitica Type III Secretion Chaperone Sycd
J.Mol.Biol., 375, 2008
|
|
4CZC
 
 | | Crystal structure of the siroheme decarboxylase NirDL in co-complex with iron-uroporphyrin III analogue | | Descriptor: | Fe(III) Uroporphyrin, NIRD-LIKE PROTEIN | | Authors: | Schmelz, S, Kriegler, T.M, Haufschildt, K, Layer, G, Heinz, D.W. | | Deposit date: | 2014-04-17 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Crystal Structure of Siroheme Decarboxylase in Complex with Iron-Uroporphyrin III Reveals Two Essential Histidine Residues
J.Mol.Biol., 426, 2014
|
|
2V5G
 
 | | Crystal structure of the mutated N263A YscU C-terminal domain | | Descriptor: | CHLORIDE ION, YSCU | | Authors: | Wiesand, U, Sorg, I, Amstutz, M, Wagner, S, Van Den Heuvel, J, Luehrs, T, Cornelis, G.R, Heinz, D.W. | | Deposit date: | 2008-10-06 | | Release date: | 2008-11-04 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Type III Secretion Recognition Protein Yscu from Yersinia Enterocolitica
J.Mol.Biol., 385, 2009
|
|
2VGY
 
 | | Crystal structure of the Yersinia enterocolitica Type III Secretion Translocator Chaperone SycD (alternative dimer) | | Descriptor: | CHAPERONE SYCD | | Authors: | Buttner, C.R, Sorg, I, Cornelis, G.R, Heinz, D.W, Niemann, H.H. | | Deposit date: | 2007-11-16 | | Release date: | 2007-12-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the Yersinia Enterocolitica Type III Secretion Chaperone Sycd
J.Mol.Biol., 375, 2008
|
|
