1B67
 
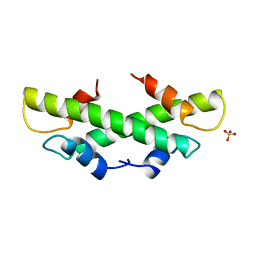 | | CRYSTAL STRUCTURE OF THE HISTONE HMFA FROM METHANOTHERMUS FERVIDUS | | 分子名称: | PROTEIN (HISTONE HMFA), SULFATE ION | | 著者 | Decanniere, K, Sandman, K, Reeve, J.N, Heinemann, U. | | 登録日 | 1999-01-19 | | 公開日 | 2000-01-17 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Crystal structures of recombinant histones HMfA and HMfB from the hyperthermophilic archaeon Methanothermus fervidus.
J.Mol.Biol., 303, 2000
|
|
1B6W
 
 | |
1CSQ
 
 | |
1CPN
 
 | |
1CSP
 
 | |
1CPM
 
 | |
466D
 
 | | DISORDER AND TWIN REFINEMENT OF RNA HEPTAMER DOUBLE HELIX | | 分子名称: | RNA (5'-R(*GP*GP*GP*GP*CP*UP*A)-3'), RNA (5'-R(*UP*AP*GP*CP*UP*CP*C)-3'), SODIUM ION, ... | | 著者 | Mueller, U, Muller, Y.A, Herbst-Irmer, R, Sprinzl, M, Heinemann, U. | | 登録日 | 1999-04-14 | | 公開日 | 1999-08-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.16 Å) | | 主引用文献 | Disorder and twin refinement of RNA heptamer double helices.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
2X6W
 
 | | Tailspike protein mutant E372Q of E.coli bacteriophage HK620 in complex with hexasaccharide | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SODIUM ION, TAIL SPIKE PROTEIN, ... | | 著者 | Lorenzen, N.K, Mueller, J.J, Heinemann, U, Seckler, R, Barbirz, S. | | 登録日 | 2010-02-22 | | 公開日 | 2011-03-02 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Single Amino Acid Exchange in Bacteriophage Hk620 Tailspike Protein Results in Thousand-Fold Increase of its Oligosaccharide Affinity.
Glycobiology, 23, 2013
|
|
2X6Y
 
 | | Tailspike protein mutant D339A of E.coli bacteriophage HK620 in complex with hexasaccharide | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SODIUM ION, TAIL SPIKE PROTEIN, ... | | 著者 | Lorenzen, N.K, Mueller, J.J, Heinemann, U, Seckler, R, Barbirz, S. | | 登録日 | 2010-02-22 | | 公開日 | 2011-03-02 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Single Amino Acid Exchange in Bacteriophage Hk620 Tailspike Protein Results in Thousand-Fold Increase of its Oligosaccharide Affinity.
Glycobiology, 23, 2013
|
|
2X85
 
 | | Tailspike protein of E. coli bacteriophage HK620 in complex with hexasaccharide | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SODIUM ION, TAILSPIKE PROTEIN HK620, ... | | 著者 | Lorenzen, N.K, Mueller, J.J, Heinemann, U, Seckler, R, Barbirz, S. | | 登録日 | 2010-03-05 | | 公開日 | 2011-03-23 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Single Amino Acid Exchange in Bacteriophage Hk620 Tailspike Protein Results in Thousand-Fold Increase of its Oligosaccharide Affinity.
Glycobiology, 23, 2013
|
|
6G8W
 
 | |
6H10
 
 | | Crystal Structure of KDM4D with tetrazolylhydrazide ligand NR073 | | 分子名称: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | 著者 | Malecki, P.H, Weiss, M.S, Heinemann, U, Link, A. | | 登録日 | 2018-07-10 | | 公開日 | 2020-01-29 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.104 Å) | | 主引用文献 | Crystal Structure of KDM4D with tetrazolylhydrazide ligand NR073
To be published
|
|
6H0Y
 
 | | Crystal Structure of KDM4D with tetrazolylhydrazide ligand NS022 | | 分子名称: | (2~{R})-3-(4-methoxyphenyl)-2-(2~{H}-1,2,3,4-tetrazol-5-yl)propanehydrazide, 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, ... | | 著者 | Malecki, P.H, Weiss, M.S, Heinemann, U, Link, A. | | 登録日 | 2018-07-10 | | 公開日 | 2020-01-29 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.212 Å) | | 主引用文献 | Crystal Structure of KDM4D with tetrazolylhydrazide ligand NS022
To be published
|
|
6H0Z
 
 | | Crystal Structure of KDM4D with tetrazolylhydrazide ligand NR067 | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysine-specific demethylase 4D, ... | | 著者 | Malecki, P.H, Weiss, M.S, Heinemann, U, Link, A. | | 登録日 | 2018-07-10 | | 公開日 | 2020-01-29 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.34 Å) | | 主引用文献 | Crystal Structure of KDM4D with tetrazolylhydrazide ligand NR067
To be published
|
|
6H0X
 
 | | Crystal Structure of KDM4D with tetrazolylhydrazide ligand AA040 | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysine-specific demethylase 4D, ... | | 著者 | Malecki, P.H, Weiss, M.S, Heinemann, U, Link, A. | | 登録日 | 2018-07-10 | | 公開日 | 2020-01-29 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Crystal Structure of KDM4D with tetrazolylhydrazide ligand AA040
To be published
|
|
6H11
 
 | | Crystal Structure of KDM4D with tetrazolylhydrazide ligand AA028 | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysine-specific demethylase 4D, ... | | 著者 | Malecki, P.H, Weiss, M.S, Heinemann, U, Link, A. | | 登録日 | 2018-07-10 | | 公開日 | 2020-01-29 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.516 Å) | | 主引用文献 | Crystal Structure of KDM4D with tetrazolylhydrazide ligand AA028
To be published
|
|
6H0W
 
 | | Crystal Structure of KDM4D with tetrazolylhydrazide ligand NS035 | | 分子名称: | (2~{R})-3-phenyl-2-(2~{H}-1,2,3,4-tetrazol-5-yl)propanehydrazide, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | 著者 | Malecki, P.H, Weiss, M.S, Heinemann, U, Link, A. | | 登録日 | 2018-07-10 | | 公開日 | 2020-01-29 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.23 Å) | | 主引用文献 | Crystal Structure of KDM4D with tetrazolylhydrazide ligand NS035
To be published
|
|
6HQC
 
 | |
3O5N
 
 | | Tetrahydroquinoline carboxylates are potent inhibitors of the Shank PDZ domain, a putative target in autism disorders | | 分子名称: | (3aS,4R,9bR)-9-nitro-3a,4,5,9b-tetrahydro-3H-cyclopenta[c]quinoline-4,6-dicarboxylic acid, SH3 and multiple ankyrin repeat domains protein 3 | | 著者 | Saupe, J, Roske, Y, Schillinger, C, Kamdem, N, Radetzki, S, Diehl, A, Oschkinat, H, Krause, G, Heinemann, U, Rademann, J. | | 登録日 | 2010-07-28 | | 公開日 | 2011-06-15 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Discovery, structure-activity relationship studies, and crystal structure of nonpeptide inhibitors bound to the shank3 PDZ domain.
Chemmedchem, 6, 2011
|
|
2XC1
 
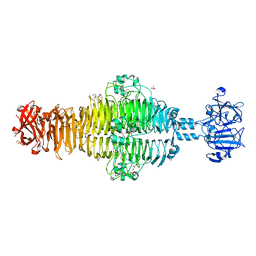 | | Full-length Tailspike Protein Mutant Y108W of Bacteriophage P22 | | 分子名称: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, BIFUNCTIONAL TAIL PROTEIN, CALCIUM ION, ... | | 著者 | Mueller, J.J, Seul, A, Seckler, R, Heinemann, U. | | 登録日 | 2010-04-15 | | 公開日 | 2011-05-04 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Bacteriophage P22 Tailspike: Structure of the Complete Protein and Function of the Interdomain Linker
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2YIL
 
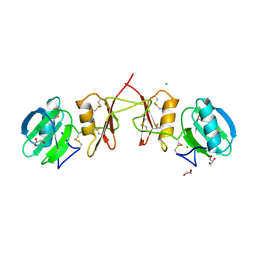 | | Crystal Structure of Parasite Sarcocystis muris Lectin SML-2 | | 分子名称: | CHLORIDE ION, GLYCEROL, MICRONEME ANTIGEN L2, ... | | 著者 | Mueller, J.J, Weiss, M.S, Heinemann, U. | | 登録日 | 2011-05-16 | | 公開日 | 2011-11-02 | | 最終更新日 | 2019-07-17 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Pan-Modular Structure of Microneme Protein Sml-2 from Parasite Sarcocystis Muris at 1.95 A Resolution and its Complex with 1-Thio-Beta-D-Galactose.
Acta Crystallogr.,Sect.D, D67, 2011
|
|
3PDO
 
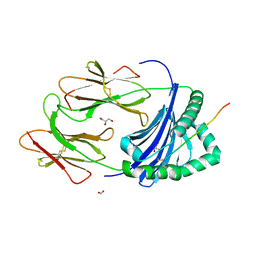 | | Crystal Structure of HLA-DR1 with CLIP102-120 | | 分子名称: | FORMIC ACID, GLYCEROL, HLA class II histocompatibility antigen gamma chain, ... | | 著者 | Gunther, S, Schlundt, A, Sticht, J, Roske, Y, Heinemann, U, Wiesmuller, K.-H, Jung, G, Falk, K, Rotzschke, O, Freund, C. | | 登録日 | 2010-10-23 | | 公開日 | 2010-12-08 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Bidirectional binding of invariant chain peptides to an MHC class II molecule.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3PF4
 
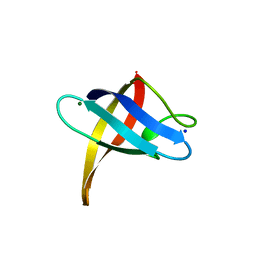 | | Crystal structure of Bs-CspB in complex with r(GUCUUUA) | | 分子名称: | Cold shock protein cspB, MAGNESIUM ION, SODIUM ION, ... | | 著者 | Sachs, R, Max, K.E.A, Heinemann, U. | | 登録日 | 2010-10-27 | | 公開日 | 2011-09-21 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | RNA single strands bind to a conserved surface of the major cold shock protein in crystals and solution.
Rna, 18, 2012
|
|
3PF5
 
 | |
2YIP
 
 | |
