2XEU
 
 | | Ring domain | | Descriptor: | RING FINGER PROTEIN 4, SULFATE ION, ZINC ION, ... | | Authors: | Plechanovova, A, McMahon, S.A, Johnson, K.A, Navratilova, I, Naismith, J.H, Hay, R.T. | | Deposit date: | 2010-05-18 | | Release date: | 2010-07-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanism of Ubiquitylation by Dimeric Ring Ligase Rnf4
Nat.Struct.Mol.Biol., 18, 2011
|
|
2XPH
 
 | | Crystal structure of human SENP1 with the bound cobalt | | Descriptor: | COBALT (II) ION, GLYCEROL, SENTRIN-SPECIFIC PROTEASE 1 | | Authors: | Rimsa, V, Eadsforth, T, Hay, R.T, Hunter, W.N. | | Deposit date: | 2010-08-26 | | Release date: | 2010-09-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Role of Co2+ in the Crystallization of Human Senp1 and Comments on the Limitations of Automated Refinement Protocols
Acta Crystallogr.,Sect.F, 67, 2011
|
|
2XRE
 
 | | Detection of cobalt in previously unassigned human SENP1 structure | | Descriptor: | COBALT (II) ION, GLYCEROL, SENTRIN-SPECIFIC PROTEASE 1 | | Authors: | Rimsa, V, Eadsforth, T, Hay, R.T, Hunter, W.N. | | Deposit date: | 2010-09-14 | | Release date: | 2010-10-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The Role of Co2+ in the Crystallization of Human Senp1 and Comments on the Limitations of Automated Refinement Protocols
Acta Crystallogr.,Sect.F, 67, 2011
|
|
2CKH
 
 | | SENP1-SUMO2 complex | | Descriptor: | SENTRIN-SPECIFIC PROTEASE 1, SMALL UBIQUITIN-RELATED MODIFIER 2 | | Authors: | Shen, L.N, Dong, C, Liu, H, Hay, R.T, Naismith, J.H. | | Deposit date: | 2006-04-18 | | Release date: | 2006-04-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Structure of Senp1-Sumo-2 Complex Suggests a Structural Basis for Discrimination between Sumo Paralogues During Processing.
Biochem.J., 397, 2006
|
|
7KIB
 
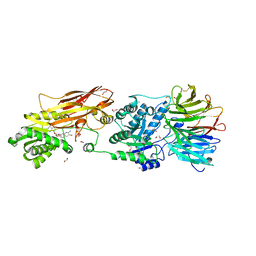 | | PRMT5:MEP50 Complexed with 5,5-Bicyclic Inhibitor Compound 4 | | Descriptor: | (2R,3R,3aS,6S,6aR)-6-[(2-amino-3-bromoquinolin-7-yl)oxy]-2-(4-amino-7H-pyrrolo[2,3-d]pyrimidin-7-yl)hexahydro-3aH-cyclopenta[b]furan-3,3a-diol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Palte, R.L, Hayes, R.P. | | Deposit date: | 2020-10-23 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | The Discovery of Two Novel Classes of 5,5-Bicyclic Nucleoside-Derived PRMT5 Inhibitors for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
5U0A
 
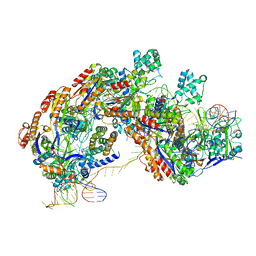 | | CRISPR RNA-guided surveillance complex | | Descriptor: | CRISPR-associated protein, Cas5e family, Cse1 family, ... | | Authors: | Xiao, Y, Luo, M, Hayes, R.P, Kim, J, Ng, S, Ding, F, Liao, M, Ke, A. | | Deposit date: | 2016-11-23 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure Basis for Directional R-loop Formation and Substrate Handover Mechanisms in Type I CRISPR-Cas System.
Cell, 170, 2017
|
|
5U07
 
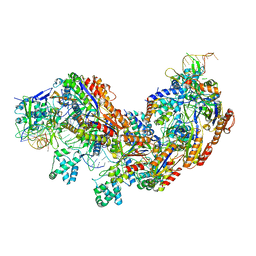 | | CRISPR RNA-guided surveillance complex | | Descriptor: | CRISPR-associated protein, Cas5e family, Cse1 family, ... | | Authors: | Xiao, Y, Luo, M, Hayes, R.P, Kim, J, Ng, S, Ding, F, Liao, M, Ke, A. | | Deposit date: | 2016-11-23 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure Basis for Directional R-loop Formation and Substrate Handover Mechanisms in Type I CRISPR-Cas System.
Cell, 170, 2017
|
|
4G0L
 
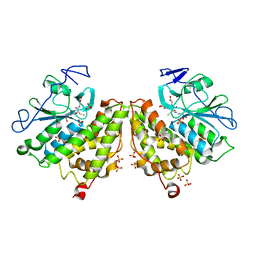 | | Glutathionyl-hydroquinone Reductase, YqjG, of E.coli complexed with GSH | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, SULFATE ION, ... | | Authors: | Green, A.R, Hayes, R.P, Xun, L, Kang, C. | | Deposit date: | 2012-07-09 | | Release date: | 2012-09-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural understanding of the glutathione-dependent reduction mechanism of glutathionyl-hydroquinone reductases.
J.Biol.Chem., 287, 2012
|
|
4G0K
 
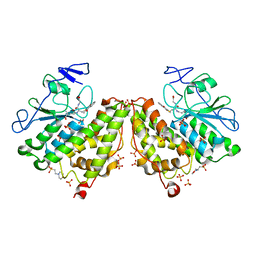 | | Glutathionyl-hydroquinone reductase, YqjG, of E.coli complexed with GS-menadione | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, L-gamma-glutamyl-S-(3-methyl-1,4-dioxo-1,4-dihydronaphthalen-2-yl)-L-cysteinylglycine, SULFATE ION, ... | | Authors: | Green, A.R, Hayes, R.P, Xun, L, Kang, C. | | Deposit date: | 2012-07-09 | | Release date: | 2012-09-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.557 Å) | | Cite: | Structural understanding of the glutathione-dependent reduction mechanism of glutathionyl-hydroquinone reductases.
J.Biol.Chem., 287, 2012
|
|
4G0I
 
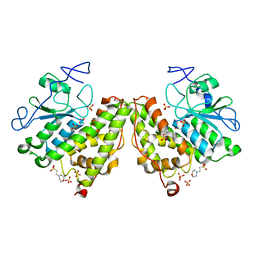 | | Glutathionyl-Hydroquinone Reductase, YqjG of Escherichia coli | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, protein yqjG | | Authors: | Green, A.R, Hayes, R.P, Xun, L, Kang, C. | | Deposit date: | 2012-07-09 | | Release date: | 2012-09-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural understanding of the glutathione-dependent reduction mechanism of glutathionyl-hydroquinone reductases.
J.Biol.Chem., 287, 2012
|
|
4FQU
 
 | |
4KEC
 
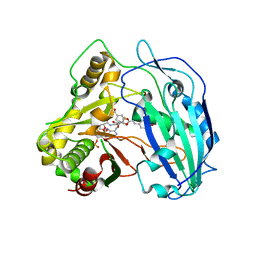 | | SbHCT-complex form | | Descriptor: | (3R,4S,5R)-3,4-dihydroxy-5-{[(2E)-3-(4-hydroxyphenyl)prop-2-enoyl]oxy}cyclohex-1-ene-1-carboxylic acid, COENZYME A, Hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyl transferase | | Authors: | Walker, A.M, Hayes, R.P, Youn, B, Vermerris, W, Sattler, S.E, Kang, C. | | Deposit date: | 2013-04-25 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Elucidation of the structure and reaction mechanism of sorghum hydroxycinnamoyltransferase and its structural relationship to other coenzyme a-dependent transferases and synthases.
Plant Physiol., 162, 2013
|
|
4KE4
 
 | | Elucidation of the structure and reaction mechanism of Sorghum bicolor hydroxycinnamoyltransferase and its structural relationship to other CoA-dependent transferases and synthases | | Descriptor: | GLYCEROL, Hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyl transferase, TETRAETHYLENE GLYCOL | | Authors: | Walker, A.M, Hayes, R.P, Youn, B, Vermerris, W, Sattler, S.E, Kang, C. | | Deposit date: | 2013-04-25 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.012 Å) | | Cite: | Elucidation of the structure and reaction mechanism of sorghum hydroxycinnamoyltransferase and its structural relationship to other coenzyme a-dependent transferases and synthases.
Plant Physiol., 162, 2013
|
|
1JQ7
 
 | | HCMV protease dimer-interface mutant, S225Y complexed to Inhibitor BILC 408 | | Descriptor: | ASSEMBLIN, N-(6-aminohexanoyl)-3-methyl-L-valyl-3-methyl-L-valyl-N~1~-[(2S,3S)-3-hydroxy-4-oxo-4-{[(1R)-1-phenylpropyl]amino}butan-2-yl]-N~4~,N~4~-dimethyl-L-aspartamide | | Authors: | Batra, R, Khayat, R, Tong, L. | | Deposit date: | 2001-08-03 | | Release date: | 2001-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular mechanism for dimerization to regulate the catalytic activity of human cytomegalovirus protease.
Nat.Struct.Biol., 8, 2001
|
|
1JQ6
 
 | |
6W6P
 
 | |
6WAX
 
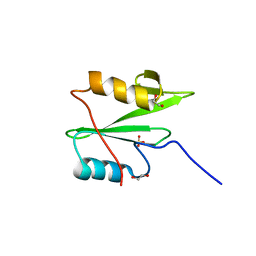 | | C-terminal SH2 domain of p120RasGAP | | Descriptor: | 1,2-ETHANEDIOL, Ras GTPase-activating protein 1, SULFATE ION | | Authors: | Jaber Chehayeb, R, Wang, J, Stiegler, A.L, Boggon, T.J. | | Deposit date: | 2020-03-26 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The GTPase-activating protein p120RasGAP has an evolutionarily conserved "FLVR-unique" SH2 domain.
J.Biol.Chem., 295, 2020
|
|
1WPX
 
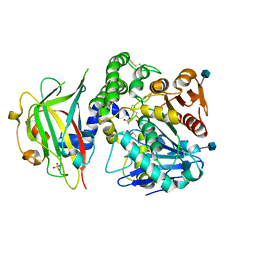 | | Crystal structure of carboxypeptidase Y inhibitor complexed with the cognate proteinase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Carboxypeptidase Y, Carboxypeptidase Y inhibitor, ... | | Authors: | Mima, J, Hayashida, M, Fujii, T, Narita, Y, Hayashi, R, Ueda, M, Hata, Y. | | Deposit date: | 2004-09-14 | | Release date: | 2005-03-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the carboxypeptidase y inhibitor i(c) in complex with the cognate proteinase reveals a novel mode of the proteinase-protein inhibitor interaction
J.Mol.Biol., 346, 2005
|
|
2OF9
 
 | |
6JKQ
 
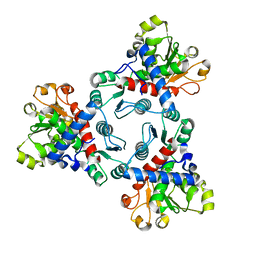 | | Crystal structure of aspartate transcarbamoylase from Trypanosoma cruzi (Ligand-free form) | | Descriptor: | Aspartate carbamoyltransferase | | Authors: | Matoba, K, Shiba, T, Nara, T, Aoki, T, Nagasaki, S, Hayamizu, R, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Balogun, E.O, Inaoka, D.K, Kita, K, Harada, S. | | Deposit date: | 2019-03-01 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystallographic snapshots of Trypanosoma cruzi aspartate transcarbamoylase
revealed an ordered Bi-Bi reaction mechanism
To Be Published
|
|
6JL6
 
 | | Crystal structure of aspartate transcarbamoylase from Trypanosoma cruzi in complex with phosphate (Pi). | | Descriptor: | ASPARTIC ACID, Aspartate carbamoyltransferase, PHOSPHATE ION | | Authors: | Matoba, K, Shiba, T, Nara, T, Aoki, T, Nagasaki, S, Hayamizu, R, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Balogun, E.O, Inaoka, D.K, Kita, K, Harada, S. | | Deposit date: | 2019-03-04 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic snapshots of Trypanosoma cruzi aspartate transcarbamoylase
revealed an ordered Bi-Bi reaction mechanism
To Be Published
|
|
6JKR
 
 | | Crystal structure of aspartate transcarbamoylase from Trypanosoma cruzi in complex with carbamoyl phosphate (CP) | | Descriptor: | Aspartate carbamoyltransferase, GLYCEROL, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER | | Authors: | Matoba, K, Shiba, T, Nara, T, Aoki, T, Nagasaki, S, Hayamizu, R, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Balogun, E.O, Inaoka, D.K, Kita, K, Harada, S. | | Deposit date: | 2019-03-01 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic snapshots of Trypanosoma cruzi aspartate transcarbamoylase
revealed an ordered Bi-Bi reaction mechanism
To Be Published
|
|
6JL4
 
 | | Crystal structure of aspartate transcarbamoylase from Trypanosoma cruzi in complex with carbamoyl aspartate (CA) and phosphate (Pi) | | Descriptor: | ASPARTIC ACID, Aspartate carbamoyltransferase, N-CARBAMOYL-L-ASPARTATE, ... | | Authors: | Matoba, K, Shiba, T, Nara, T, Aoki, T, Nagasaki, S, Hayamizu, R, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Balogun, E.O, Inaoka, D.K, Kita, K, Harada, S. | | Deposit date: | 2019-03-04 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic snapshots of Trypanosoma cruzi aspartate transcarbamoylase
revealed an ordered Bi-Bi reaction mechanism
To Be Published
|
|
6JKS
 
 | | Crystal structure of aspartate transcarbamoylase from Trypanosoma cruzi in complex with carbamoyl phosphate (CP) and aspartate (Asp) | | Descriptor: | ASPARTIC ACID, Aspartate carbamoyltransferase, putative, ... | | Authors: | Matoba, K, Shiba, T, Nara, T, Aoki, T, Nagasaki, S, Hayamizu, R, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Balogun, E.O, Inaoka, D.K, Kita, K, Harada, S. | | Deposit date: | 2019-03-01 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic snapshots of Trypanosoma cruzi aspartate transcarbamoylase
revealed an ordered Bi-Bi reaction mechanism
To Be Published
|
|
6JL5
 
 | | Crystal structure of aspartate transcarbamoylase from Trypanosoma cruzi in complex with aspartate (Asp) and phosphate (Pi). | | Descriptor: | ASPARTIC ACID, Aspartate carbamoyltransferase, GLYCEROL, ... | | Authors: | Matoba, K, Shiba, T, Nara, T, Aoki, T, Nagasaki, S, Hayamizu, R, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Balogun, E.O, Inaoka, D.K, Kita, K, Harada, S. | | Deposit date: | 2019-03-04 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystallographic snapshots of Trypanosoma cruzi aspartate transcarbamoylase
revealed an ordered Bi-Bi reaction mechanism
To Be Published
|
|
