1Y3Q
 
 | | Structure of AlgQ1, alginate-binding protein | | Descriptor: | AlgQ1, CALCIUM ION | | Authors: | Momma, K, Mishima, Y, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2004-11-26 | | Release date: | 2005-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Direct Evidence for Sphingomonas sp. A1 Periplasmic Proteins as Macromolecule-Binding Proteins Associated with the ABC Transporter: Molecular Insights into Alginate Transport in the Periplasm(,)
Biochemistry, 44, 2005
|
|
1Y3I
 
 | | Crystal Structure of Mycobacterium tuberculosis NAD kinase-NAD complex | | Descriptor: | GLYCEROL, Inorganic polyphosphate/ATP-NAD kinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Mori, S, Yamasaki, M, Maruyama, Y, Momma, K, Kawai, S, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2004-11-25 | | Release date: | 2005-01-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | NAD-binding mode and the significance of intersubunit contact revealed by the crystal structure of Mycobacterium tuberculosis NAD kinase-NAD complex
Biochem.Biophys.Res.Commun., 327, 2005
|
|
1Y3P
 
 | | Structure of AlgQ1, alginate-binding protein, complexed with an alginate tetrasaccharide | | Descriptor: | AlgQ1, CALCIUM ION, beta-D-mannopyranuronic acid-(1-4)-alpha-D-mannopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-D-mannopyranuronic acid | | Authors: | Momma, K, Mishima, Y, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2004-11-26 | | Release date: | 2005-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Direct Evidence for Sphingomonas sp. A1 Periplasmic Proteins as Macromolecule-Binding Proteins Associated with the ABC Transporter: Molecular Insights into Alginate Transport in the Periplasm(,)
Biochemistry, 44, 2005
|
|
1Y3N
 
 | | Structure of AlgQ1, alginate-binding protein, complexed with an alginate disaccharide | | Descriptor: | AlgQ1, CALCIUM ION, beta-D-mannopyranuronic acid-(1-4)-alpha-D-mannopyranuronic acid | | Authors: | Momma, K, Mishima, Y, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2004-11-26 | | Release date: | 2005-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Direct Evidence for Sphingomonas sp. A1 Periplasmic Proteins as Macromolecule-Binding Proteins Associated with the ABC Transporter: Molecular Insights into Alginate Transport in the Periplasm(,)
Biochemistry, 44, 2005
|
|
1Y3H
 
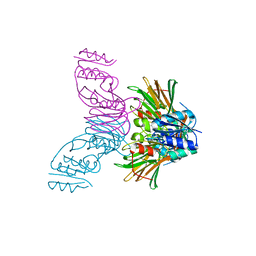 | | Crystal Structure of Inorganic Polyphosphate/ATP-NAD kinase from Mycobacterium tuberculosis | | Descriptor: | Inorganic polyphosphate/ATP-NAD kinase | | Authors: | Mori, S, Yamasaki, M, Maruyama, Y, Momma, K, kawai, S, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2004-11-24 | | Release date: | 2005-01-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | NAD-binding mode and the significance of intersubunit contact revealed by the crystal structure of Mycobacterium tuberculosis NAD kinase-NAD complex
BIOCHEM.BIOPHYS.RES.COMMUN., 327, 2005
|
|
6INZ
 
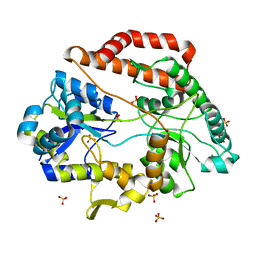 | | Crystal structure of solute-binding protein complexed with unsaturated hyaluronan disaccharide | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Extracellular solute-binding protein family 1, ... | | Authors: | Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2018-10-29 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.289 Å) | | Cite: | Substrate recognition by bacterial solute-binding protein is responsible for import of extracellular hyaluronan and chondroitin sulfate from the animal host.
Biosci.Biotechnol.Biochem., 83, 2019
|
|
6JHX
 
 | |
5XS8
 
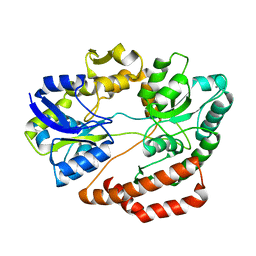 | | Crystal structure of solute-binding protein complexed with unsaturated chondroitin disaccharide with two sulfate groups at C-4 and C-6 positions of GalNAc | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4,6-di-O-sulfo-beta-D-galactopyranose, CALCIUM ION, Extracellular solute-binding protein family 1 | | Authors: | Oiki, S, Kamochi, R, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2017-06-12 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Alternative substrate-bound conformation of bacterial solute-binding protein involved in the import of mammalian host glycosaminoglycans.
Sci Rep, 7, 2017
|
|
5Z6C
 
 | |
5Z6B
 
 | | Crystal structure of sugar-binding protein YesO in complex with rhamnogalacturonan trisaccharide | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-2)-[beta-D-galactopyranose-(1-4)]alpha-L-rhamnopyranose, Putative ABC transporter substrate-binding protein YesO | | Authors: | Sugiura, H, Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2018-01-22 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.582 Å) | | Cite: | Crystal structure of sugar-binding protein YesO in complex with rhamnogalacturonan trisaccharide
To Be Published
|
|
5Y4C
 
 | | Crystal structure of EfeO-like protein Algp7 in complex with a metal ion | | Descriptor: | Alginate-binding protein, COPPER (II) ION, GLYCEROL | | Authors: | Temtrirath, K, Maruyama, Y, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2017-08-03 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Binding mode of metal ions to the bacterial iron import protein EfeO
Biochem. Biophys. Res. Commun., 493, 2017
|
|
6A6U
 
 | | Crystal structure of the modified fructosyl peptide oxidase from Aspergillus nidulans with R61G mutation, in complex with FSA | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, 1-S-(carboxymethyl)-1-thio-beta-D-fructopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Ogawa, N, Maruyama, Y, Itoh, T, Hashimoto, W, Murata, K. | | Deposit date: | 2018-06-29 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.945 Å) | | Cite: | Creation of haemoglobin A1c direct oxidase from fructosyl peptide oxidase by combined structure-based site specific mutagenesis and random mutagenesis.
Sci Rep, 9, 2019
|
|
6A6T
 
 | | Crystal structure of the modified fructosyl peptide oxidase from Aspergillus nidulans with R61G mutation | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl amine: oxygen oxidoreductase, ... | | Authors: | Ogawa, N, Maruyama, Y, Itoh, T, Hashimoto, W, Murata, K. | | Deposit date: | 2018-06-29 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Creation of haemoglobin A1c direct oxidase from fructosyl peptide oxidase by combined structure-based site specific mutagenesis and random mutagenesis.
Sci Rep, 9, 2019
|
|
6A6V
 
 | | Crystal structure of the modified fructosyl peptide oxidase from Aspergillus nidulans with 7 additional mutations, in complex with FSA | | Descriptor: | 1-S-(carboxymethyl)-1-thio-beta-D-fructopyranose, FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl amine: oxygen oxidoreductase | | Authors: | Ogawa, N, Maruyama, Y, Itoh, T, Hashimoto, W, Murata, K. | | Deposit date: | 2018-06-29 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Creation of haemoglobin A1c direct oxidase from fructosyl peptide oxidase by combined structure-based site specific mutagenesis and random mutagenesis.
Sci Rep, 9, 2019
|
|
6A6R
 
 | | Crystal structure of the modified fructosyl peptide oxidase from Aspergillus nidulans, Seleno-methionine Derivative | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl amine: oxygen oxidoreductase, ... | | Authors: | Ogawa, N, Maruyama, Y, Itoh, T, Hashimoto, W, Murata, K. | | Deposit date: | 2018-06-29 | | Release date: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | Creation of haemoglobin A1c direct oxidase from fructosyl peptide oxidase by combined structure-based site specific mutagenesis and random mutagenesis.
Sci Rep, 9, 2019
|
|
6A6S
 
 | | Crystal structure of the modified fructosyl peptide oxidase from Aspergillus nidulans in complex with FSA, Seleno-methionine Derivative | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, 1-S-(carboxymethyl)-1-thio-beta-D-fructopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Ogawa, N, Maruyama, Y, Itoh, T, Hashimoto, W, Murata, K. | | Deposit date: | 2018-06-29 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Creation of haemoglobin A1c direct oxidase from fructosyl peptide oxidase by combined structure-based site specific mutagenesis and random mutagenesis.
Sci Rep, 9, 2019
|
|
2D8L
 
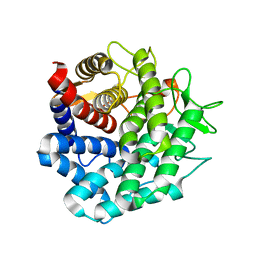 | | Crystal Structure of Unsaturated Rhamnogalacturonyl Hydrolase in complex with dGlcA-GalNAc | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose, Putative glycosyl hydrolase yteR | | Authors: | Itoh, T, Ochiai, A, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2005-12-06 | | Release date: | 2006-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A novel glycoside hydrolase family 105: the structure of family 105 unsaturated rhamnogalacturonyl hydrolase complexed with a disaccharide in comparison with family 88 enzyme complexed with the disaccharide
J.Mol.Biol., 360, 2006
|
|
2AHG
 
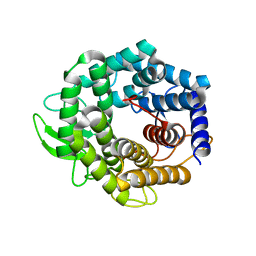 | | Unsaturated glucuronyl hydrolase mutant D88N with dGlcA-GalNAc | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose, unsaturated glucuronyl hydrolase | | Authors: | Itoh, T, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2005-07-28 | | Release date: | 2006-08-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Unsaturated Glucuronyl Hydrolase Complexed with Substrate: MOLECULAR INSIGHTS INTO ITS CATALYTIC REACTION MECHANISM
J.Biol.Chem., 281, 2006
|
|
2D5J
 
 | |
2E24
 
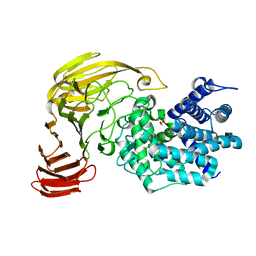 | | crystal structure of a mutant (R612A) of xanthan lyase | | Descriptor: | DI(HYDROXYETHYL)ETHER, Xanthan lyase | | Authors: | Maruyama, Y, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2006-11-07 | | Release date: | 2007-01-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A Structural Factor Responsible for Substrate Recognition by Bacillus sp. GL1 Xanthan Lyase that Acts Specifically on Pyruvated Side Chains of Xanthan
Biochemistry, 46, 2007
|
|
2CWS
 
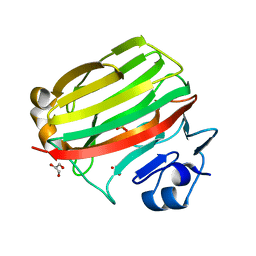 | | Crystal structure at 1.0 A of alginate lyase A1-II', a member of polysaccharide lyase family-7 | | Descriptor: | GLYCEROL, SULFATE ION, alginate lyase A1-II' | | Authors: | Yamasaki, M, Ogura, K, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2005-06-25 | | Release date: | 2005-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A Structural Basis for Depolymerization of Alginate by Polysaccharide Lyase Family-7
J.Mol.Biol., 352, 2005
|
|
2E22
 
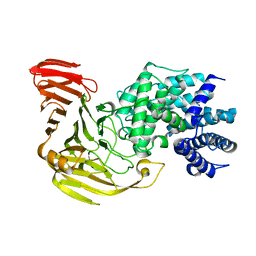 | | Crystal structure of xanthan lyase in complex with mannose | | Descriptor: | Xanthan lyase, alpha-D-mannopyranose | | Authors: | Maruyama, Y, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2006-11-07 | | Release date: | 2007-01-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Structural Factor Responsible for Substrate Recognition by Bacillus sp. GL1 Xanthan Lyase that Acts Specifically on Pyruvated Side Chains of Xanthan
Biochemistry, 46, 2007
|
|
