1J0N
 
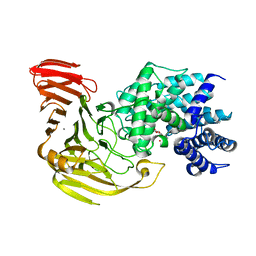 | | Crystal Structure of Bacillus sp. GL1 Xanthan Lyase that Acts on Side Chains of Xanthan | | Descriptor: | 4,6-O-[(1S)-1-carboxyethylidene]-beta-D-glucopyranose, CALCIUM ION, XANTHAN LYASE | | Authors: | Hashimoto, W, Nankai, H, Mikami, B, Murata, K. | | Deposit date: | 2002-11-19 | | Release date: | 2003-04-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Bacillus sp. GL1 Xanthan Lyase, Which Acts on the Side Chains of Xanthan.
J.Biol.Chem., 278, 2003
|
|
1J0M
 
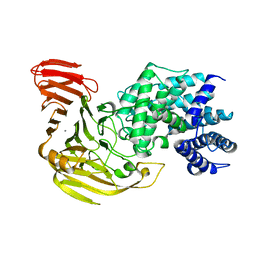 | | Crystal Structure of Bacillus sp. GL1 Xanthan Lyase that Acts on Side Chains of Xanthan | | Descriptor: | CALCIUM ION, XANTHAN LYASE | | Authors: | Hashimoto, W, Nankai, H, Mikami, B, Murata, K. | | Deposit date: | 2002-11-19 | | Release date: | 2003-04-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Bacillus sp. GL1 Xanthan Lyase, Which Acts on the Side Chains of Xanthan.
J.Biol.Chem., 278, 2003
|
|
4U8E
 
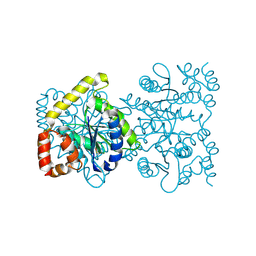 | | Crystal structure of 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase from Streptococcus agalactiae | | Descriptor: | Putative uncharacterized protein gbs1892 | | Authors: | Maruyama, y, Oiki, S, Takase, R, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2014-08-03 | | Release date: | 2015-01-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metabolic Fate of Unsaturated Glucuronic/Iduronic Acids from Glycosaminoglycans: MOLECULAR IDENTIFICATION AND STRUCTURE DETERMINATION OF STREPTOCOCCAL ISOMERASE AND DEHYDROGENASE
J.Biol.Chem., 290, 2015
|
|
3K3T
 
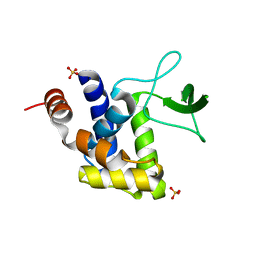 | | E185A mutant of peptidoglycan hydrolase from Sphingomonas sp. A1 | | Descriptor: | Peptidoglycan hydrolase FlgJ, SULFATE ION | | Authors: | Maruyama, Y, Ochiai, A, Itoh, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2009-10-04 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mutational studies of the peptidoglycan hydrolase FlgJ of Sphingomonas sp. strain A1
J.Basic Microbiol., 50, 2010
|
|
3K8V
 
 | |
2OKX
 
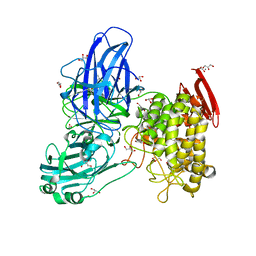 | | Crystal structure of GH78 family rhamnosidase of Bacillus SP. GL1 AT 1.9 A | | Descriptor: | CALCIUM ION, GLYCEROL, Rhamnosidase B | | Authors: | Cui, Z, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2007-01-17 | | Release date: | 2007-11-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Glycoside Hydrolase Family 78 alpha-L-Rhamnosidase from Bacillus sp. GL1
J.Mol.Biol., 374, 2007
|
|
4W7H
 
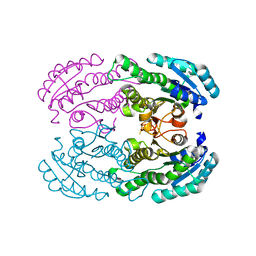 | | Crystal Structure of DEH Reductase A1-R Mutant | | Descriptor: | Carbonyl reductase | | Authors: | Takase, R, Mikami, B, Kawai, S, Murata, K, Hashimoto, W. | | Deposit date: | 2014-08-22 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structure-based Conversion of the Coenzyme Requirement of a Short-chain Dehydrogenase/Reductase Involved in Bacterial Alginate Metabolism.
J.Biol.Chem., 289, 2014
|
|
4W7I
 
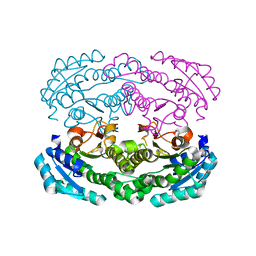 | | Crystal structure of DEH reductase A1-R' mutant | | Descriptor: | 4-deoxy-L-erythro-5-hexoseulose uronate reductase A1-R' | | Authors: | Takase, R, Mikami, B, Kawai, S, Murata, K, Hashimoto, W. | | Deposit date: | 2014-08-22 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure-based Conversion of the Coenzyme Requirement of a Short-chain Dehydrogenase/Reductase Involved in Bacterial Alginate Metabolism.
J.Biol.Chem., 289, 2014
|
|
2RGK
 
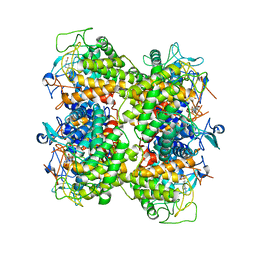 | | Functional annotation of Escherichia coli yihS-encoded protein | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Uncharacterized sugar isomerase yihS | | Authors: | Itoh, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2007-10-03 | | Release date: | 2008-08-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of YihS in complex with D-mannose: structural annotation of Escherichia coli and Salmonella enterica yihS-encoded proteins to an aldose-ketose isomerase
J.Mol.Biol., 377, 2008
|
|
1KWH
 
 | | Structure Analysis AlgQ2, a Macromolecule(alginate)-Binding Periplasmic Protein of Sphingomonas sp. A1. | | Descriptor: | CALCIUM ION, Macromolecule-Binding Periplasmic Protein | | Authors: | Momma, K, Mikami, B, Mishima, Y, Hashimoto, W, Murata, K. | | Deposit date: | 2002-01-29 | | Release date: | 2002-02-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of AlgQ2, a macromolecule (alginate)-binding protein of Sphingomonas sp. A1 at 2.0A resolution.
J.Mol.Biol., 316, 2002
|
|
8YRI
 
 | | Crystal structure of sugar phosphotransferase system EIIB component CPF_0401 from Clostridium perfringens | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PTS system, mannose/fructose/sorbose family, ... | | Authors: | Hirayama, Y, Oiki, S, Mikami, B, Ogura, K, Hashimoto, W. | | Deposit date: | 2024-03-21 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of sugar phosphotransferase system EIIB component CPF_0401 from Clostridium perfringens
To Be Published
|
|
7YE3
 
 | | Crystal structure of Lactobacillus rhamnosus 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase KduI complexed with MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase, ZINC ION | | Authors: | Yamamoto, Y, Oiki, S, Takase, R, Mikami, B, Hashimoto, W. | | Deposit date: | 2022-07-05 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.553 Å) | | Cite: | Crystal Structures of Lacticaseibacillus 4-Deoxy-L- threo- 5-hexosulose-uronate Ketol-isomerase KduI in Complex with Substrate Analogs.
J Appl Glycosci (1999), 70, 2023
|
|
7YRS
 
 | | Crystal structure of Lactobacillus rhamnosus 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase KduI complexed with MOPS | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase, ZINC ION | | Authors: | Yamamoto, Y, Oiki, S, Takase, R, Mikami, B, Hashimoto, W. | | Deposit date: | 2022-08-10 | | Release date: | 2023-08-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Crystal Structures of Lacticaseibacillus 4-Deoxy-L- threo- 5-hexosulose-uronate Ketol-isomerase KduI in Complex with Substrate Analogs.
J Appl Glycosci (1999), 70, 2023
|
|
4MMH
 
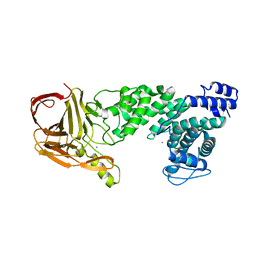 | | Crystal structure of heparan sulfate lyase HepC from Pedobacter heparinus | | Descriptor: | CALCIUM ION, Heparinase III protein | | Authors: | Maruyama, Y, Nakamichi, Y, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2013-09-09 | | Release date: | 2014-01-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Pedobacter heparinus Heparin Lyase Hep III with the Active Site in a Deep Cleft
Biochemistry, 53, 2014
|
|
4MMI
 
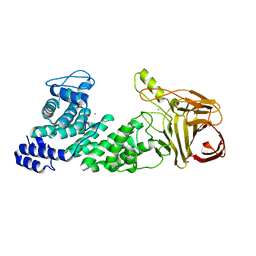 | | Crystal structure of heparan sulfate lyase HepC mutant from Pedobacter heparinus | | Descriptor: | CALCIUM ION, Heparinase III protein | | Authors: | Maruyama, Y, Nakamichi, Y, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2013-09-09 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Pedobacter heparinus Heparin Lyase Hep III with the Active Site in a Deep Cleft
Biochemistry, 53, 2014
|
|
3IM0
 
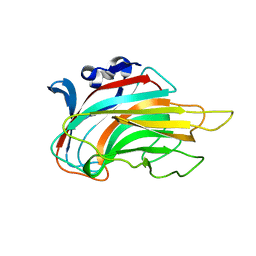 | | Crystal structure of Chlorella virus vAL-1 soaked in 200mM D-glucuronic acid, 10% PEG-3350, and 200mM glycine-NaOH (pH 10.0) | | Descriptor: | VAL-1, beta-D-glucopyranuronic acid | | Authors: | Ogura, K, Yamasaki, M, Hashidume, T, Yamada, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2009-08-08 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of family 14 polysaccharide lyase with pH-dependent modes of action
J.Biol.Chem., 284, 2009
|
|
3GNE
 
 | | Crystal structure of alginate lyase vAL-1 from Chlorella virus | | Descriptor: | CITRATE ANION, GLYCEROL, VAL-1 | | Authors: | Ogura, K, Yamasaki, M, Hashidume, T, Yamada, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2009-03-17 | | Release date: | 2009-10-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of family 14 polysaccharide lyase with pH-dependent modes of action
J.Biol.Chem., 284, 2009
|
|
3K8W
 
 | |
1FP3
 
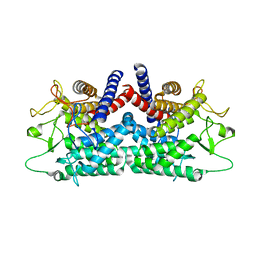 | | CRYSTAL STRUCTURE OF N-ACYL-D-GLUCOSAMINE 2-EPIMERASE FROM PORCINE KIDNEY | | Descriptor: | N-ACYL-D-GLUCOSAMINE 2-EPIMERASE | | Authors: | Itoh, T, Mikami, B, Maru, I, Ohta, Y, Hashimoto, W, Murata, K. | | Deposit date: | 2000-08-30 | | Release date: | 2000-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of N-acyl-D-glucosamine 2-epimerase from porcine kidney at 2.0 A resolution.
J.Mol.Biol., 303, 2000
|
|
7WGU
 
 | | Crystal structure of metal-binding protein EfeO from Escherichia coli | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Iron uptake system protein EfeO, ... | | Authors: | Nakatsuji, S, Takase, R, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-12-29 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of EfeB and EfeO in a bacterial siderophore-independent iron transport system
Biochem.Biophys.Res.Commun., 594, 2022
|
|
2Z8R
 
 | | Crystal structure of rhamnogalacturonan lyase YesW at 1.40 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, YesW protein | | Authors: | Ochiai, A, Itoh, T, Maruyama, Y, Kawamata, A, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2007-09-10 | | Release date: | 2007-10-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Novel Structural Fold in Polysaccharide Lyases: BACILLUS SUBTILIS FAMILY 11 RHAMNOGALACTURONAN LYASE YesW WITH AN EIGHT-BLADED -PROPELLER
J.Biol.Chem., 282, 2007
|
|
7E4S
 
 | | Crystal structure of Lactobacillus rhamnosus 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase KduI complexed with HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-dehydro-4-deoxy-D-glucuronate isomerase, ZINC ION | | Authors: | Yamamoto, Y, Takase, R, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-02-15 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal structures of Lacticaseibacillus 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase KduI in complex with substrate analogs
J.Appl.Glyosci., 2023
|
|
2ZUX
 
 | | Crystal structure of rhamnogalacturonan lyase YesW complexed with rhamnose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, YesW protein, ... | | Authors: | Ochiai, A, Itoh, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2008-10-28 | | Release date: | 2009-02-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural determinants responsible for substrate recognition and mode of action in family 11 polysaccharide lyases
J.Biol.Chem., 284, 2009
|
|
7VEQ
 
 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in an open conformation | | Descriptor: | GLYCEROL, SPH1118 | | Authors: | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-09-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
2Z8S
 
 | | Crystal structure of rhamnogalacturonan lyase YesW complexed with digalacturonic acid | | Descriptor: | CALCIUM ION, YesW protein, alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid | | Authors: | Ochiai, A, Itoh, T, Maruyama, Y, Kawamata, A, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2007-09-10 | | Release date: | 2007-10-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Novel Structural Fold in Polysaccharide Lyases: BACILLUS SUBTILIS FAMILY 11 RHAMNOGALACTURONAN LYASE YesW WITH AN EIGHT-BLADED -PROPELLER
J.Biol.Chem., 282, 2007
|
|
