2I3S
 
 | | Bub3 complex with Bub1 GLEBS motif | | Descriptor: | Cell cycle arrest protein, Checkpoint serine/threonine-protein kinase | | Authors: | Larsen, N.A, Harrison, S.C. | | Deposit date: | 2006-08-20 | | Release date: | 2007-01-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of Bub3 interactions in the mitotic spindle checkpoint.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3N4X
 
 | | Structure of Csm1 full-length | | Descriptor: | Monopolin complex subunit CSM1 | | Authors: | Corbett, K.D, Harrison, S.C. | | Deposit date: | 2010-05-23 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.408 Å) | | Cite: | The Monopolin Complex Crosslinks Kinetochore Components to Regulate Chromosome-Microtubule Attachments.
Cell(Cambridge,Mass.), 142, 2010
|
|
1XI4
 
 | | Clathrin D6 Coat | | Descriptor: | Clathrin heavy chain, Clathrin light chain A | | Authors: | Fotin, A, Cheng, Y, Sliz, P, Grigorieff, N, Harrison, S.C, Kirchhausen, T, Walz, T. | | Deposit date: | 2004-09-21 | | Release date: | 2004-11-02 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Molecular model for a complete clathrin lattice from electron cryomicroscopy
Nature, 432, 2004
|
|
1XI5
 
 | | Clathrin D6 coat with auxilin J-domain | | Descriptor: | Auxilin J-domain, Clathrin heavy chain | | Authors: | Fotin, A, Cheng, Y, Grigorieff, N, Walz, T, Harrison, S.C, Kirchhausen, T. | | Deposit date: | 2004-09-21 | | Release date: | 2004-11-02 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structure of an auxilin-bound clathrin coat and its implications for the mechanism of uncoating
Nature, 432, 2004
|
|
3N4S
 
 | | Structure of Csm1 C-terminal domain, P21212 form | | Descriptor: | Monopolin complex subunit CSM1, PENTAETHYLENE GLYCOL | | Authors: | Corbett, K.D, Harrison, S.C. | | Deposit date: | 2010-05-22 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Monopolin Complex Crosslinks Kinetochore Components to Regulate Chromosome-Microtubule Attachments.
Cell(Cambridge,Mass.), 142, 2010
|
|
3ZXA
 
 | |
2OF3
 
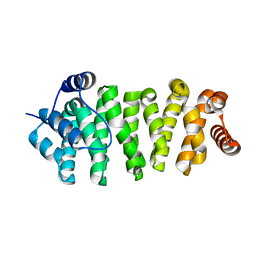 | |
3N7N
 
 | | Structure of Csm1/Lrs4 complex | | Descriptor: | Monopolin complex subunit CSM1, Monopolin complex subunit LRS4 | | Authors: | Corbett, K.D, Harrison, S.C. | | Deposit date: | 2010-05-27 | | Release date: | 2010-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | The Monopolin Complex Crosslinks Kinetochore Components to Regulate Chromosome-Microtubule Attachments.
Cell(Cambridge,Mass.), 142, 2010
|
|
3N4R
 
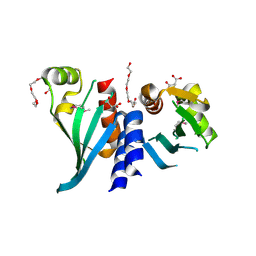 | | Structure of Csm1 C-terminal domain, R3 form | | Descriptor: | MALONATE ION, Monopolin complex subunit CSM1, PENTAETHYLENE GLYCOL | | Authors: | Corbett, K.D, Harrison, S.C. | | Deposit date: | 2010-05-22 | | Release date: | 2010-09-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | The Monopolin Complex Crosslinks Kinetochore Components to Regulate Chromosome-Microtubule Attachments.
Cell(Cambridge,Mass.), 142, 2010
|
|
1PBW
 
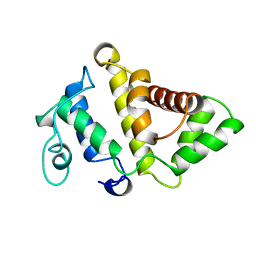 | | STRUCTURE OF BCR-HOMOLOGY (BH) DOMAIN | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE | | Authors: | Musacchio, A, Cantley, L.C, Harrison, S.C. | | Deposit date: | 1996-10-17 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the breakpoint cluster region-homology domain from phosphoinositide 3-kinase p85 alpha subunit.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1SIE
 
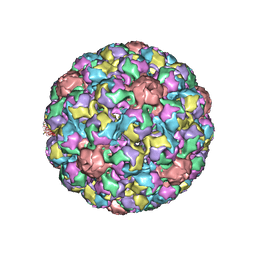 | | MURINE POLYOMAVIRUS COMPLEXED WITH A DISIALYLATED OLIGOSACCHARIDE | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-[N-acetyl-alpha-neuraminic acid-(2-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, POLYOMAVIRUS COAT PROTEIN VP1 | | Authors: | Stehle, T, Harrison, S.C. | | Deposit date: | 1995-12-12 | | Release date: | 1996-06-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Crystal structures of murine polyomavirus in complex with straight-chain and branched-chain sialyloligosaccharide receptor fragments.
Structure, 4, 1996
|
|
6CBJ
 
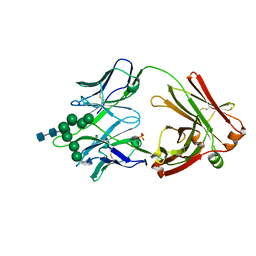 | | Crystal Structure of DH270.3 Fab in complex with Man9 | | Descriptor: | DH270.3 Fab heavy chain, DH270.3 Fab light chain, PHOSPHATE ION, ... | | Authors: | Fera, D, Harrison, S.C. | | Deposit date: | 2018-02-03 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | HIV envelope V3 region mimic embodies key features of a broadly neutralizing antibody lineage epitope.
Nat Commun, 9, 2018
|
|
6CBP
 
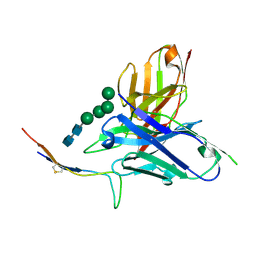 | |
6P7W
 
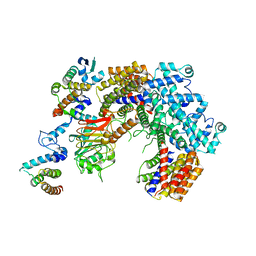 | | Structure of the K. lactis CBF3 core - Ndc10 D1 complex | | Descriptor: | Cep3, Ctf13, Ndc10, ... | | Authors: | Lee, P.D, Wei, H, Tan, D, Harrison, S.C. | | Deposit date: | 2019-06-06 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of the Centromere Binding Factor 3 Complex from Kluyveromyces lactis.
J.Mol.Biol., 431, 2019
|
|
6P7V
 
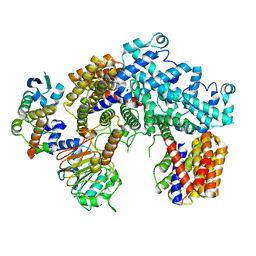 | | Structure of the K. lactis CBF3 core | | Descriptor: | Cep3, Ctf13, Skp1 | | Authors: | Lee, P.D, Wei, H, Tan, D, Harrison, S.C. | | Deposit date: | 2019-06-06 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of the Centromere Binding Factor 3 Complex from Kluyveromyces lactis.
J.Mol.Biol., 431, 2019
|
|
6P7X
 
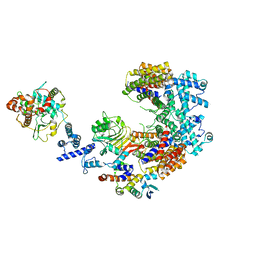 | | Structure of the K. lactis CBF3 core - Ndc10 D1D2 complex | | Descriptor: | Cep3, Ctf13, Ndc10, ... | | Authors: | Lee, P.D, Wei, H, Tan, D, Harrison, S.C. | | Deposit date: | 2019-06-06 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure of the Centromere Binding Factor 3 Complex from Kluyveromyces lactis.
J.Mol.Biol., 431, 2019
|
|
1NFI
 
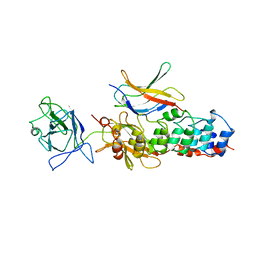 | | I-KAPPA-B-ALPHA/NF-KAPPA-B COMPLEX | | Descriptor: | I-KAPPA-B-ALPHA, NF-KAPPA-B P50, NF-KAPPA-B P65 | | Authors: | Jacobs, M.D, Harrison, S.C. | | Deposit date: | 1998-08-25 | | Release date: | 1998-11-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of an IkappaBalpha/NF-kappaB complex.
Cell(Cambridge,Mass.), 95, 1998
|
|
1YSA
 
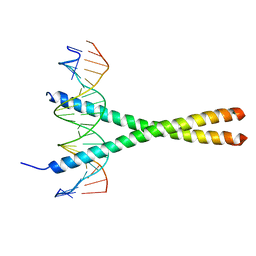 | | THE GCN4 BASIC REGION LEUCINE ZIPPER BINDS DNA AS A DIMER OF UNINTERRUPTED ALPHA HELICES: CRYSTAL STRUCTURE OF THE PROTEIN-DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*AP*CP*TP*GP*GP*AP*TP*GP*AP*GP*TP*CP*AP*TP*A P*GP*GP*A)-3'), DNA (5'-D(*TP*TP*CP*CP*TP*AP*TP*GP*AP*CP*TP*CP*AP*TP*CP*CP*A P*GP*TP*T)-3'), PROTEIN (GCN4) | | Authors: | Ellenberger, T.E, Brandl, C.J, Struhl, K, Harrison, S.C. | | Deposit date: | 1993-08-09 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: crystal structure of the protein-DNA complex.
Cell(Cambridge,Mass.), 71, 1992
|
|
6Q0E
 
 | |
6CFZ
 
 | |
1N1H
 
 | | Initiation complex of polymerase lambda3 from reovirus | | Descriptor: | 3'-DEOXY-CYTIDINE-5'-TRIPHOSPHATE, 3'-DEOXY-GUANOSINE-5'-TRIPHOSPHATE, 5'-R(*AP*UP*UP*AP*GP*C)-3', ... | | Authors: | Tao, Y, Farsetta, D.L, Nibert, M.L, Harrison, S.C. | | Deposit date: | 2002-10-17 | | Release date: | 2002-12-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | RNA Synthesis in a Cage--Structural Studies of Reovirus Polymerase [lambda] 3
Cell(Cambridge,Mass.), 111, 2002
|
|
4AU6
 
 | | Location of the dsRNA-dependent polymerase, VP1, in rotavirus particles | | Descriptor: | RNA-DEPENDENT RNA POLYMERASE | | Authors: | Estrozi, L.F, Settembre, E.C, Goret, G, McClain, B, Zhang, X, Chen, J.Z, Grigorieff, N, Harrison, S.C. | | Deposit date: | 2012-05-14 | | Release date: | 2012-06-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Location of the Dsrna-Dependent Polymerase, Vp1, in Rotavirus Particles.
J.Mol.Biol., 425, 2013
|
|
2AJF
 
 | | Structure of SARS coronavirus spike receptor-binding domain complexed with its receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme-Related Carboxypeptidase (Ace2), CHLORIDE ION, ... | | Authors: | Li, F, Li, W, Farzan, M, Harrison, S.C. | | Deposit date: | 2005-08-01 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of SARS coronavirus spike receptor-binding domain complexed with receptor.
Science, 309, 2005
|
|
5TRP
 
 | |
5U0R
 
 | |
