1OGL
 
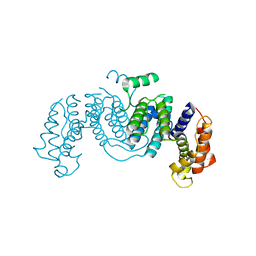 | | The crystal structure of native Trypanosoma cruzi dUTPase | | Descriptor: | DEOXYURIDINE TRIPHOSPHATASE | | Authors: | Harkiolaki, M, Dodson, E.J, Bernier-Villamor, V, Turkenburg, J.P, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2003-05-07 | | Release date: | 2004-01-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of Trypanosoma Cruzi Dutpase Reveals a Novel Dutp/Dudp Binding Fold
Structure, 12, 2004
|
|
1OEB
 
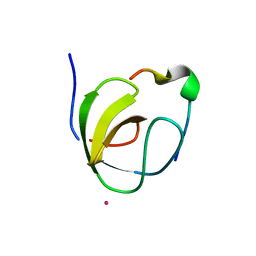 | | Mona/Gads SH3C domain | | Descriptor: | CADMIUM ION, GRB2-RELATED ADAPTOR PROTEIN 2, LYMPHOCYTE CYTOSOLIC PROTEIN 2 | | Authors: | Harkiolaki, M, Lewitzky, M, Gilbert, R.J.C, Jones, E.Y, Bourette, R.P, Mouchiroud, G, Sondermann, H, Moarefi, I, Feller, S.M. | | Deposit date: | 2003-03-24 | | Release date: | 2003-04-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Basis for SH3 Domain-Mediated High-Affinity Binding between Mona/Gads and Slp-76
Embo J., 22, 2003
|
|
1OGK
 
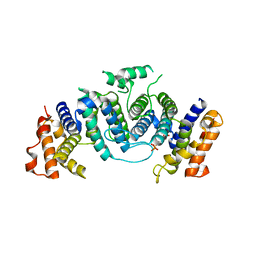 | | The crystal structure of Trypanosoma cruzi dUTPase in complex with dUDP | | Descriptor: | DEOXYURIDINE TRIPHOSPHATASE, DEOXYURIDINE-5'-DIPHOSPHATE | | Authors: | Harkiolaki, M, Dodson, E.J, Bernier-Villamor, V, Turkenburg, J.P, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2003-05-07 | | Release date: | 2004-01-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The Crystal Structure of Trypanosoma Cruzi Dutpase Reveals a Novel Dutp/Dudp Binding Fold
Structure, 12, 2004
|
|
2BZX
 
 | |
2WBJ
 
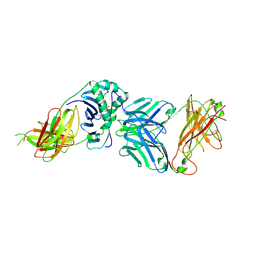 | | TCR complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HLA CLASS II HISTOCOMPATIBILITY ANTIGEN, DR ALPHA CHAIN, ... | | Authors: | Harkiolaki, M, Holmes, S.L, Svendsen, P, Gregersen, J.W, Jensen, L.T, McMahon, R, Friese, M.A, van Boxel, G, Etzensperger, R, Tzartos, J.S, Kranc, K, Sainsbury, S, Harlos, K, Mellins, E.D, Palace, J, Esiri, M.M, van der Merwe, P.A, Jones, E.Y, Fugger, L. | | Deposit date: | 2009-03-02 | | Release date: | 2009-04-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | T Cell-Mediated Autoimmune Disease due to Low-Affinity Crossreactivity to Common Microbial Peptides.
Immunity, 30, 2009
|
|
2BZY
 
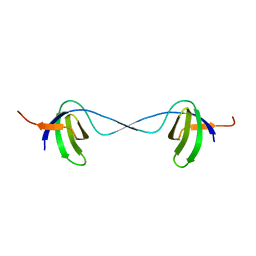 | |
2W10
 
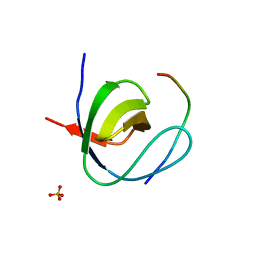 | | Mona SH3C in complex | | Descriptor: | GRB2-RELATED ADAPTOR PROTEIN 2, PHOSPHATE ION, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 23 | | Authors: | Harkiolaki, M, Feller, S.M. | | Deposit date: | 2008-10-13 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Distinct Binding Modes of Two Epitopes in Gab2 that Interact with the Sh3C Domain of Grb2.
Structure, 17, 2009
|
|
2W0Z
 
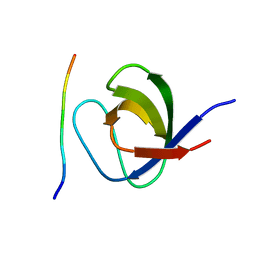 | | Grb2 SH3C (3) | | Descriptor: | GRB2-ASSOCIATED-BINDING PROTEIN 2, GROWTH FACTOR RECEPTOR-BOUND PROTEIN 2 | | Authors: | Harkiolaki, M, Tsirka, T, Feller, S.M. | | Deposit date: | 2008-10-13 | | Release date: | 2009-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Distinct Binding Modes of Two Epitopes in Gab2 that Interact with the Sh3C Domain of Grb2.
Structure, 17, 2009
|
|
2VWF
 
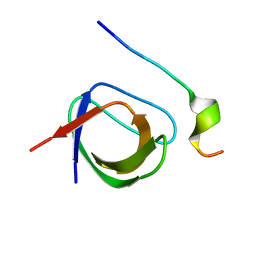 | | Grb2 SH3C (2) | | Descriptor: | GRB2-ASSOCIATED-BINDING PROTEIN 2, GROWTH FACTOR RECEPTOR-BOUND PROTEIN 2 | | Authors: | Harkiolaki, M, Tsirka, T, Feller, S.M. | | Deposit date: | 2008-06-24 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Distinct Binding Modes of Two Epitopes in Gab2 that Interact with the Sh3C Domain of Grb2.
Structure, 17, 2009
|
|
2VVK
 
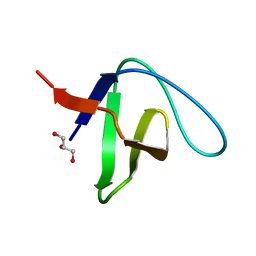 | | Grb2 SH3C (1) | | Descriptor: | GLYCEROL, GROWTH FACTOR RECEPTOR-BOUND PROTEIN 2 | | Authors: | Harkiolaki, M, Tsirka, T, Feller, S.M. | | Deposit date: | 2008-06-09 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Distinct Binding Modes of Two Epitopes in Gab2 that Interact with the Sh3C Domain of Grb2.
Structure, 17, 2009
|
|
1UTI
 
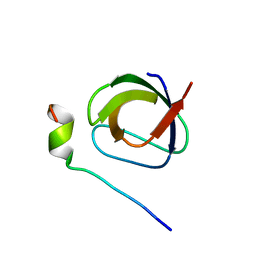 | | Mona/Gads SH3C in complex with HPK derived peptide | | Descriptor: | GRB2-RELATED ADAPTOR PROTEIN 2, MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE KINASE 1 | | Authors: | Lewitzky, M, Harkiolaki, M, Domart, M.C, Feller, S.M. | | Deposit date: | 2003-12-09 | | Release date: | 2004-05-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mona/Gads Sh3C Binding to Hematopoietic Progenitor Kinase 1 (Hpk1) Combines an Atypical SH3 Binding Motif, R/Kxxk, with a Classical Pxxp Motif Embedded in a Polyproline Type II (Ppii) Helix
J.Biol.Chem., 279, 2004
|
|
1W2Y
 
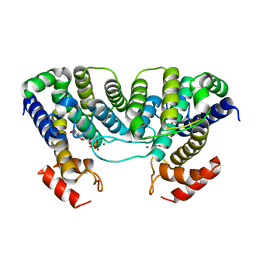 | | The crystal structure of a complex of Campylobacter jejuni dUTPase with substrate analogue dUpNHp | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-DIPHOSPHATE, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDE HYDROLASE, MAGNESIUM ION | | Authors: | Moroz, O.V, Harkiolaki, M, Galperin, M.Y, Vagin, A.A, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2004-07-09 | | Release date: | 2004-09-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Crystal Structure of a Complex of Campylobacter Jejuni Dutpase with Substrate Analogue Sheds Light on the Mechanism and Suggests the "Basic Module" for Dimeric D(C/U)Tpases
J.Mol.Biol., 342, 2004
|
|
2J63
 
 | | Crystal structure of AP endonuclease LMAP from Leishmania major | | Descriptor: | AP-ENDONUCLEASE | | Authors: | Vidal, A.E, Harkiolaki, M, Gallego, C, Castillo-Acosta, V.M, Ruiz-Perez, L.M, Wilson, K.S, Gonzalez-Pacanowska, D. | | Deposit date: | 2006-09-25 | | Release date: | 2007-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal Structure and DNA Repair Activities of the Ap Endonuclease from Leishmania Major.
J.Mol.Biol., 373, 2007
|
|
6VQ2
 
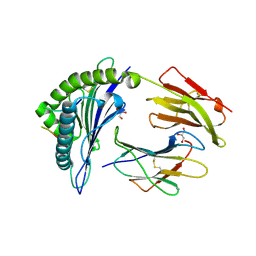 | | HLA-B*27:05 presenting an HIV-1 14mer peptide | | Descriptor: | 14-mer peptide, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | Deposit date: | 2020-02-04 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
6VQE
 
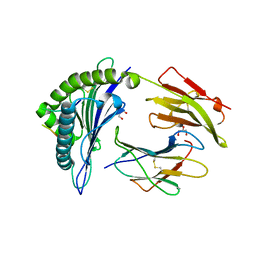 | | HLA-B*27:05 presenting an HIV-1 13mer peptide | | Descriptor: | 13-mer peptide, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | Deposit date: | 2020-02-05 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
6VQZ
 
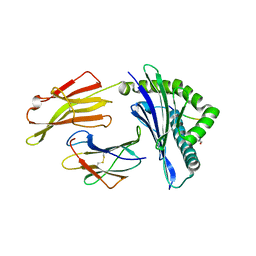 | | HLA-B*27:05 presenting an HIV-1 6mer peptide | | Descriptor: | 6-mer peptide, ARGININE, Beta-2-microglobulin, ... | | Authors: | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | Deposit date: | 2020-02-06 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
6VPZ
 
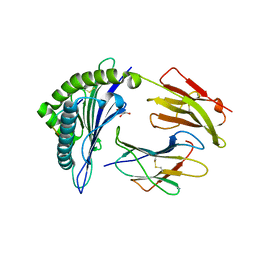 | | HLA-B*27:05 presenting an HIV-1 11mer peptide | | Descriptor: | 11-mer peptide, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | Deposit date: | 2020-02-04 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
6VQD
 
 | | HLA-B*27:05 presenting an HIV-1 8mer peptide | | Descriptor: | 8-mer peptide, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | Deposit date: | 2020-02-05 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
6VQY
 
 | | HLA-B*27:05 presenting an HIV-1 7mer peptide | | Descriptor: | 7-mer peptide, ARGININE, Beta-2-microglobulin, ... | | Authors: | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | Deposit date: | 2020-02-06 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
2XX6
 
 | | Structure of the Bacillus subtilis prophage dUTPase, YosS | | Descriptor: | SPBC2 PROPHAGE-DERIVED DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE YOSS | | Authors: | Garcia-Nafria, J, Harkiolaki, M, Persson, R, Fogg, M.J, Wilson, K.S. | | Deposit date: | 2010-11-08 | | Release date: | 2011-02-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The Structure of Bacillus Subtilis Sp Beta Prophage Dutpase and its Complexes with Two Nucleotides
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2XY3
 
 | | Structure of the Bacillus subtilis prophage dUTPase with dUpNHpp | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, MAGNESIUM ION, SPBC2 PROPHAGE-DERIVED DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE YOSS | | Authors: | Garcia-Nafria, J, Harkiolaki, M, Persson, R, Fogg, M.J, Wilson, K.S. | | Deposit date: | 2010-11-12 | | Release date: | 2011-02-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The Structure of Bacillus Subtilis Sp Beta Prophage Dutpase and its Complexes with Two Nucleotides
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2Y1T
 
 | | Bacillus subtilis prophage dUTPase in complex with dUDP | | Descriptor: | DEOXYURIDINE-5'-DIPHOSPHATE, SPBC2 PROPHAGE-DERIVED DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE YOSS | | Authors: | Garcia-Nafria, J, Harkiolaki, M, Persson, R, Fogg, M.J, Wilson, K.S. | | Deposit date: | 2010-12-10 | | Release date: | 2011-02-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The Structure of Bacillus Subtilis Sp Beta Prophage Dutpase and its Complexes with Two Nucleotides
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2XFX
 
 | | cattle MHC class I N01301 presenting an 11mer from Theileria parva | | Descriptor: | BETA-2-MICROGLOBULIN, MHC CLASS 1, UNCHARACTERIZED PROTEIN | | Authors: | Macdonald, I.K, Harkiolaki, M, Hunt, L, Morrison, W.I, Connelley, T, Graham, S.P, Jones, E.Y, Flower, D.R, Ellis, S.A. | | Deposit date: | 2010-05-28 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mhc Class I Bound to an Immunodominant Theileria Parva Epitope Demonstrates Unconventional Presentation to T Cell Receptors.
Plos Pathog., 6, 2010
|
|
2CIK
 
 | | Insights Into Crossreactivity in Human Allorecognition: The Structure of HLA-B35011 Presenting an Epitope derived from Cytochrome P450. | | Descriptor: | BETA-2-MICROGLOBULIN, GLYCEROL, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN B-35 ALPHA CHAIN, ... | | Authors: | Hourigan, C.S, Harkiolaki, M, Peterson, N.A, Bell, J.I, Jones, E.Y, O'Callaghan, C.A. | | Deposit date: | 2006-03-22 | | Release date: | 2006-10-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Structure of the Human Allo-Ligand Hla-B3501 in Complex with a Cytochrome P450 Peptide: Steric Hindrance Influences Tcr Allo-Recognition.
Eur.J.Immunol., 36, 2006
|
|
2CIC
 
 | | THE CRYSTAL STRUCTURE OF A COMPLEX OF CAMPYLOBACTER JEJUNI DUTPASE WITH SUBSTRATE ANALOGUE DUPNHPP | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDE HYDROLASE, MAGNESIUM ION | | Authors: | Moroz, O.V, Harkiolaki, M, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2006-03-17 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of the Leishmania Major Deoxyuridine Triphosphate Nucleotidohydrolase in Complex with Nucleotide Analogues, Dump, and Deoxyuridine.
J.Biol.Chem., 286, 2011
|
|
