2KZI
 
 | | Solution structure of the ZHER2 Affibody | | 分子名称: | Engineered protein, ZHER2 Affibody | | 著者 | Hard, T. | | 登録日 | 2010-06-18 | | 公開日 | 2010-08-04 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for high-affinity HER2 receptor binding by an engineered protein.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2KZJ
 
 | |
2M5A
 
 | |
1UTR
 
 | | UTEROGLOBIN-PCB COMPLEX (REDUCED FORM) | | 分子名称: | 4,4'-BIS([H]METHYLSULFONYL)-2,2',5,5'-TETRACHLOROBIPHENYL, UTEROGLOBIN | | 著者 | Hard, T, Barnes, H.J, Larsson, C, Gustafsson, J.-A, Lund, J. | | 登録日 | 1995-09-01 | | 公開日 | 1995-12-07 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a mammalian PCB-binding protein in complex with a PCB.
Nat.Struct.Biol., 2, 1995
|
|
1DFE
 
 | | NMR STRUCTURE OF RIBOSOMAL PROTEIN L36 FROM THERMUS THERMOPHILUS | | 分子名称: | L36 RIBOSOMAL PROTEIN, ZINC ION | | 著者 | Hard, T, Rak, A, Allard, P, Kloo, L, Garber, M. | | 登録日 | 1999-11-19 | | 公開日 | 1999-12-01 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of ribosomal protein L36 from Thermus thermophilus reveals a zinc-ribbon-like fold.
J.Mol.Biol., 296, 2000
|
|
1DGZ
 
 | | RIBOSMAL PROTEIN L36 FROM THERMUS THERMOPHILUS: NMR STRUCTURE ENSEMBLE | | 分子名称: | PROTEIN (L36 RIBOSOMAL PROTEIN), ZINC ION | | 著者 | Hard, T, Rak, A, Allard, P, Kloo, L, Garber, M. | | 登録日 | 1999-11-27 | | 公開日 | 1999-12-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of ribosomal protein L36 from Thermus thermophilus reveals a zinc-ribbon-like fold.
J.Mol.Biol., 296, 2000
|
|
2POG
 
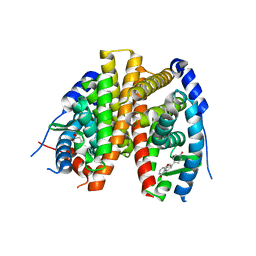 | | Benzopyrans as Selective Estrogen Receptor b Agonists (SERBAs). Part 2: Structure Activity Relationship Studies on the Benzopyran Scaffold. | | 分子名称: | (3AS,4R,9BR)-4-(4-HYDROXYPHENYL)-1,2,3,3A,4,9B-HEXAHYDROCYCLOPENTA[C]CHROMEN-9-OL, Estrogen receptor | | 著者 | Richardson, T.I, Norman, B.H, Lugar, C.W, Jones, S.A, Wang, Y, Durbin, J.D, Krishnan, V, Dodge, J.A. | | 登録日 | 2007-04-26 | | 公開日 | 2007-09-11 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Benzopyrans as selective estrogen receptor beta agonists (SERBAs). Part 2: structure-activity relationship studies on the benzopyran scaffold.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2QTU
 
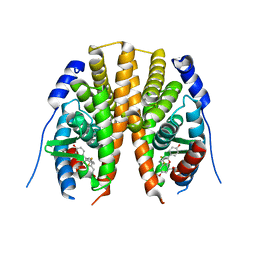 | | Estrogen receptor beta ligand-binding domain complexed to a benzopyran ligand | | 分子名称: | (3aS,4R,9bR)-2,2-difluoro-4-(4-hydroxyphenyl)-6-(methoxymethyl)-1,2,3,3a,4,9b-hexahydrocyclopenta[c]chromen-8-ol, Estrogen receptor beta | | 著者 | Richardson, T.I, Dodge, J.A, Wang, Y, Durbin, J.D, Krishnan, V, Norman, B.H. | | 登録日 | 2007-08-02 | | 公開日 | 2007-10-30 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.53 Å) | | 主引用文献 | Benzopyrans as selective estrogen receptor beta agonists (SERBAs). Part 5: Combined A- and C-ring structure-activity relationship studies.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2GDA
 
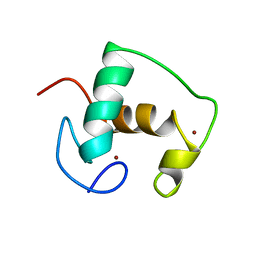 | | REFINED SOLUTION STRUCTURE OF THE GLUCOCORTICOID RECEPTOR DNA-BINDING DOMAIN | | 分子名称: | GLUCOCORTICOID RECEPTOR, ZINC ION | | 著者 | Baumann, H, Paulsen, K, Kovacs, H, Berglund, H, Wright, A.P.H, Gustafsson, J.-A, Hard, T. | | 登録日 | 1994-03-15 | | 公開日 | 1994-06-22 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Refined solution structure of the glucocorticoid receptor DNA-binding domain.
Biochemistry, 32, 1993
|
|
1BBX
 
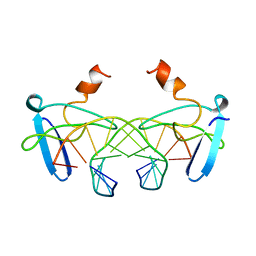 | | NON-SPECIFIC PROTEIN-DNA INTERACTIONS IN THE SSO7D-DNA COMPLEX, NMR, 1 STRUCTURE | | 分子名称: | DNA (5'-D(*CP*TP*AP*GP*CP*GP*CP*GP*CP*TP*AP*G)-3'), DNA-BINDING PROTEIN 7D | | 著者 | Agback, P, Baumann, H, Knapp, S, Ladenstein, R, Hard, T. | | 登録日 | 1998-04-24 | | 公開日 | 1998-10-14 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Architecture of Nonspecific Protein-DNA Interactions in the Sso7D-DNA Complex
Nat.Struct.Biol., 5, 1998
|
|
1GDC
 
 | | REFINED SOLUTION STRUCTURE OF THE GLUCOCORTICOID RECEPTOR DNA-BINDING DOMAIN | | 分子名称: | GLUCOCORTICOID RECEPTOR, ZINC ION | | 著者 | Baumann, H, Paulsen, K, Kovacs, H, Berglund, H, Wright, A.P.H, Gustafsson, J.-A, Hard, T. | | 登録日 | 1994-03-15 | | 公開日 | 1994-06-22 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Refined solution structure of the glucocorticoid receptor DNA-binding domain.
Biochemistry, 32, 1993
|
|
1SSO
 
 | | SOLUTION STRUCTURE AND DNA-BINDING PROPERTIES OF A THERMOSTABLE PROTEIN FROM THE ARCHAEON SULFOLOBUS SOLFATARICUS | | 分子名称: | SSO7D | | 著者 | Baumann, H, Knapp, S, Lundback, T, Ladenstein, R, Hard, T. | | 登録日 | 1995-03-31 | | 公開日 | 1995-05-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure and DNA-binding properties of a thermostable protein from the archaeon Sulfolobus solfataricus.
Nat.Struct.Biol., 1, 1994
|
|
1EAO
 
 | | THE RUNX1 Runt domain at 1.4A resolution: a structural switch and specifically bound chloride ions modulate DNA binding | | 分子名称: | BROMIDE ION, RUNT-RELATED TRANSCRIPTION FACTOR 1 | | 著者 | Backstrom, S, Wolf-Watz, M, Grundstrom, C, Hard, T.H, Grundstrom, T, Sauer, U.H. | | 登録日 | 2001-07-14 | | 公開日 | 2002-09-12 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | The Runx1 Runt Domain at 1.25A Resolution: A Structural Switch and Specifically Bound Chloride Ions Modulate DNA Binding
J.Mol.Biol., 322, 2002
|
|
1AWW
 
 | | SH3 DOMAIN FROM BRUTON'S TYROSINE KINASE, NMR, 42 STRUCTURES | | 分子名称: | BRUTON'S TYROSINE KINASE | | 著者 | Hansson, H, Mattsson, P.T, Allard, P, Haapaniemi, P, Vihinen, M, Smith, C.I.E, Hard, T. | | 登録日 | 1997-10-06 | | 公開日 | 1998-04-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the SH3 domain from Bruton's tyrosine kinase.
Biochemistry, 37, 1998
|
|
1AWX
 
 | | SH3 DOMAIN FROM BRUTON'S TYROSINE KINASE, NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | BRUTON'S TYROSINE KINASE | | 著者 | Hansson, H, Mattsson, P.T, Allard, P, Haapaniemi, P, Vihinen, M, Smith, C.I.E, Hard, T. | | 登録日 | 1997-10-06 | | 公開日 | 1998-04-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the SH3 domain from Bruton's tyrosine kinase.
Biochemistry, 37, 1998
|
|
2OTK
 
 | |
1QKH
 
 | | SOLUTION STRUCTURE OF THE RIBOSOMAL PROTEIN S19 FROM THERMUS THERMOPHILUS | | 分子名称: | 30S RIBOSOMAL PROTEIN S19 | | 著者 | Helgstrand, M, Rak, A.V, Allard, P, Davydova, N, Garber, M.B, Hard, T. | | 登録日 | 1999-07-20 | | 公開日 | 1999-07-21 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the ribosomal protein S19 from Thermus thermophilus.
J. Mol. Biol., 292, 1999
|
|
1ILF
 
 | |
2ACM
 
 | |
1N88
 
 | | NMR structure of the ribosomal protein L23 from Thermus thermophilus. | | 分子名称: | Ribosomal protein L23 | | 著者 | Ohman, A, Rak, A, Dontsova, M, Garber, M.B, Hard, T. | | 登録日 | 2002-11-20 | | 公開日 | 2003-06-10 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of the ribosomal protein L23 from Thermus thermophilus.
J.Biomol.NMR, 26, 2003
|
|
1QKF
 
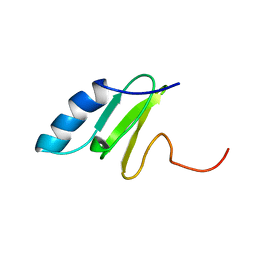 | | SOLUTION STRUCTURE OF THE RIBOSOMAL PROTEIN S19 FROM THERMUS THERMOPHILUS | | 分子名称: | 30S RIBOSOMAL PROTEIN S19 | | 著者 | Helgstrand, M, Rak, A.V, Allard, P, Davydova, N, Garber, M.B, Hard, T. | | 登録日 | 1999-07-19 | | 公開日 | 1999-07-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the ribosomal protein S19 from Thermus thermophilus.
J. Mol. Biol., 292, 1999
|
|
2VRG
 
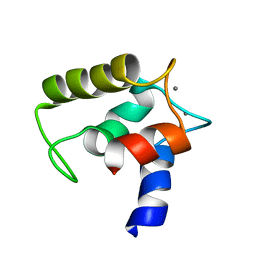 | | Structure of human MCFD2 | | 分子名称: | CALCIUM ION, MULTIPLE COAGULATION FACTOR DEFICIENCY PROTEIN 2 | | 著者 | Guy, J.E, Wigren, E, Svard, M, Hard, T, Lindqvist, Y. | | 登録日 | 2008-04-04 | | 公開日 | 2008-07-15 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | New insights into multiple coagulation factor deficiency from the solution structure of human MCFD2.
J. Mol. Biol., 381, 2008
|
|
2XET
 
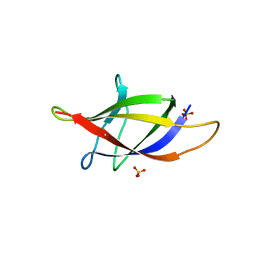 | | Conserved hydrophobic clusters on the surface of the Caf1A usher C-terminal domain are important for F1 antigen assembly | | 分子名称: | F1 CAPSULE-ANCHORING PROTEIN, SULFATE ION | | 著者 | Dubnovitsky, A.P, Duck, Z, Kersley, J.E, Hard, T, MacIntyre, S, Knight, S.D. | | 登録日 | 2010-05-17 | | 公開日 | 2010-09-22 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Conserved Hydrophobic Clusters on the Surface of the Caf1A Usher C-Terminal Domain are Important for F1 Antigen Assembly.
J.Mol.Biol., 403, 2010
|
|
1ILY
 
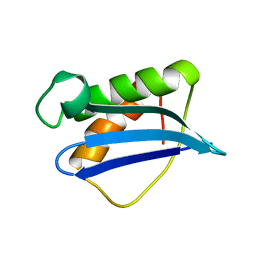 | | Solution Structure of Ribosomal Protein L18 of Thermus thermophilus | | 分子名称: | RIBOSOMAL PROTEIN L18 | | 著者 | Woestenenk, E.A, Gongadze, G.M, Shcherbakov, D.V, Rak, A.V, Garber, M.B, Hard, T, Berglund, H. | | 登録日 | 2001-05-09 | | 公開日 | 2002-05-01 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of ribosomal protein L18 from Thermus thermophilus reveals a conserved RNA-binding fold.
Biochem.J., 363, 2002
|
|
1EAN
 
 | | THE RUNX1 Runt domain at 1.70A resolution: A structural switch and specifically bound chloride ions modulate DNA binding | | 分子名称: | CHLORIDE ION, RUNT-RELATED TRANSCRIPTION FACTOR 1 | | 著者 | Backstrom, S, Wolf-Watz, M, Grundstrom, C, Hard, T, Grundstrom, T, Sauer, U.H. | | 登録日 | 2001-07-13 | | 公開日 | 2002-09-12 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The Runx1 Runt Domain at 1.25 A Resolution: A Structural Switch and Specifically Bound Chloride Ions Modulate DNA Binding
J.Mol.Biol., 322, 2002
|
|
