3IH9
 
 | |
3IHB
 
 | |
3WY8
 
 | | Crystal Structure of Protease Anisep from Arthrobacter Nicotinovorans | | 分子名称: | Serine protease | | 著者 | Sone, T, Haraguchi, Y, Kuwahara, A, Ose, T, Takano, M, Abe, A, Tanaka, M, Tanaka, I, Asano, K. | | 登録日 | 2014-08-20 | | 公開日 | 2015-08-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural characterization reveals the keratinolytic activity of an arthrobacter nicotinovorans protease.
Protein Pept.Lett., 22, 2015
|
|
3WWH
 
 | | Crystal structure of the first R-stereoselective -transaminase identified from Arthrobacter sp. KNK168 (FERM-BP-5228) | | 分子名称: | (R)-amine transaminase, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Guan, L.J, Ohtsuka, J, Okai, M, Miyakawa, T, Mase, T, Zhi, Y, Ito, N, Yasohara, Y, Tanokura, M. | | 登録日 | 2014-06-18 | | 公開日 | 2015-08-12 | | 最終更新日 | 2018-11-21 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | A new target region for changing the substrate specificity of amine transaminases.
Sci Rep, 5, 2015
|
|
3WWI
 
 | | Crystal structure of the G136F mutant of the first R-stereoselective -transaminase identified from Arthrobacter sp. KNK168 (FERM-BP-5228) | | 分子名称: | (R)-amine transaminase, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Guan, L.J, Ohtsuka, J, Miyakawa, T, Zhi, Y, Ito, N, Yasohara, Y, Tanokura, M. | | 登録日 | 2014-06-18 | | 公開日 | 2015-08-19 | | 最終更新日 | 2020-01-22 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | A new target region for changing the substrate specificity of amine transaminases.
Sci Rep, 5, 2015
|
|
3WWJ
 
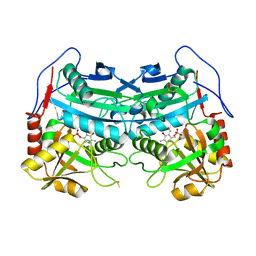 | | Crystal structure of an engineered sitagliptin-producing transaminase, ATA-117-Rd11 | | 分子名称: | (R)-amine transaminase, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Guan, L.J, Ohtsuka, J, Okai, M, Miyakawa, T, Mase, T, Zhi, Y, Hou, F, Ito, N, Yasohara, Y, Tanokura, M. | | 登録日 | 2014-06-18 | | 公開日 | 2015-08-12 | | 最終更新日 | 2018-11-21 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | A new target region for changing the substrate specificity of amine transaminases.
Sci Rep, 5, 2015
|
|
2YVE
 
 | | Crystal structure of the methylene blue-bound form of the multi-drug binding transcriptional repressor CgmR | | 分子名称: | 3,7-BIS(DIMETHYLAMINO)PHENOTHIAZIN-5-IUM, CHLORIDE ION, GLYCEROL, ... | | 著者 | Itou, H, Shirakihara, Y, Tanaka, I. | | 登録日 | 2007-04-12 | | 公開日 | 2008-04-15 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal Structures of the Multidrug Binding Repressor Corynebacteriumglutamicum CgmR in Complex with Inducers and with an Operator
J.Mol.Biol., 403, 2010
|
|
2YVH
 
 | |
2ZOZ
 
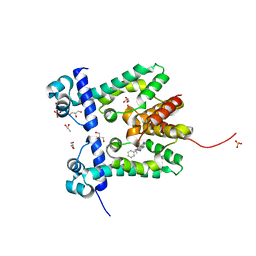 | | Crystal structure of the ethidium-bound form of the multi-drug binding transcriptional repressor CgmR | | 分子名称: | ETHIDIUM, GLYCEROL, SULFATE ION, ... | | 著者 | Itou, H, Shirakihara, Y, Tanaka, I. | | 登録日 | 2008-06-20 | | 公開日 | 2008-07-08 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal Structures of the Multidrug Binding Repressor Corynebacteriumglutamicum CgmR in Complex with Inducers and with an Operator
J.Mol.Biol., 403, 2010
|
|
2Z5H
 
 | | Crystal structure of the head-to-tail junction of tropomyosin complexed with a fragment of TnT | | 分子名称: | General control protein GCN4 and Tropomyosin alpha-1 chain, Tropomyosin alpha-1 chain and General control protein GCN4, Troponin T, ... | | 著者 | Murakami, K, Nozawa, K, Tomii, K, Kudou, N, Igarashi, N, Shirakihara, Y, Wakatsuki, S, Stewart, M, Yasunaga, T, Wakabayashi, T. | | 登録日 | 2007-07-12 | | 公開日 | 2008-04-22 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | Structural basis for tropomyosin overlap in thin (actin) filaments and the generation of a molecular swivel by troponin-T
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2Z5I
 
 | | Crystal structure of the head-to-tail junction of tropomyosin | | 分子名称: | General control protein GCN4 and Tropomyosin alpha-1 chain, MAGNESIUM ION, Tropomyosin alpha-1 chain and General control protein GCN4 | | 著者 | Murakami, K, Nozawa, K, Tomii, K, Kudou, N, Igarashi, N, Shirakihara, Y, Wakatsuki, S, Stewart, M, Yasunaga, T, Wakabayashi, T. | | 登録日 | 2007-07-12 | | 公開日 | 2008-04-22 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis for tropomyosin overlap in thin (actin) filaments and the generation of a molecular swivel by troponin-T
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2ZCT
 
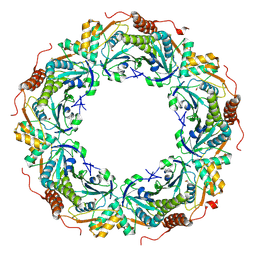 | | Oxidation of archaeal peroxiredoxin involves a hypervalent sulfur intermediate | | 分子名称: | Probable peroxiredoxin | | 著者 | Nakamura, T, Hagihara, Y, Abe, M, Inoue, T, Yamamoto, T, Matsumura, H. | | 登録日 | 2007-11-12 | | 公開日 | 2008-05-27 | | 最終更新日 | 2021-11-10 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Oxidation of archaeal peroxiredoxin involves a hypervalent sulfur intermediate
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3VW4
 
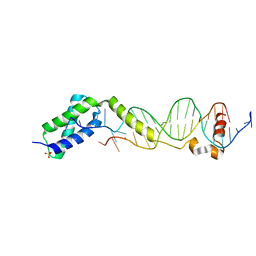 | | Crystal structure of the DNA-binding domain of ColE2-P9 Rep in complex with the replication origin | | 分子名称: | DNA (5'-D(P*AP*AP*TP*GP*AP*GP*AP*CP*CP*AP*GP*AP*TP*AP*AP*GP*CP*CP*TP*TP*AP*TP*C)-3'), DNA (5'-D(P*GP*AP*TP*AP*AP*GP*GP*CP*TP*TP*AP*TP*CP*TP*GP*GP*TP*CP*TP*CP*AP*TP*T)-3'), Rep, ... | | 著者 | Itou, H, Yagura, M, Itoh, T, Shirakihara, Y. | | 登録日 | 2012-07-31 | | 公開日 | 2013-07-31 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural Basis for Replication Origin Unwinding by An Initiator-Primase of Plasmid ColE2-P9: Duplex DNA Unwinding by A Single Protein
J.Biol.Chem., 290, 2015
|
|
3AGD
 
 | |
3WV5
 
 | | Complex structure of VinN with 3-methylaspartate | | 分子名称: | (2S,3S)-3-methyl-aspartic acid, Non-ribosomal peptide synthetase | | 著者 | Miyanaga, A, Cieslak, J, Shinohara, Y, Kudo, F, Eguchi, T. | | 登録日 | 2014-05-15 | | 公開日 | 2014-10-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The crystal structure of the adenylation enzyme VinN reveals a unique beta-amino acid recognition mechanism
J.Biol.Chem., 289, 2014
|
|
3WV4
 
 | | Crystal structure of VinN | | 分子名称: | Non-ribosomal peptide synthetase | | 著者 | Miyanaga, A, Cieslak, J, Shinohara, Y, Kudo, F, Eguchi, T. | | 登録日 | 2014-05-15 | | 公開日 | 2014-10-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | The crystal structure of the adenylation enzyme VinN reveals a unique beta-amino acid recognition mechanism
J.Biol.Chem., 289, 2014
|
|
3WVN
 
 | | Complex structure of VinN with L-aspartate | | 分子名称: | ASPARTIC ACID, Non-ribosomal peptide synthetase | | 著者 | Miyanaga, A, Cieslak, J, Shinohara, Y, Kudo, F, Eguchi, T. | | 登録日 | 2014-05-30 | | 公開日 | 2014-10-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The crystal structure of the adenylation enzyme VinN reveals a unique beta-amino acid recognition mechanism
J.Biol.Chem., 289, 2014
|
|
3WI3
 
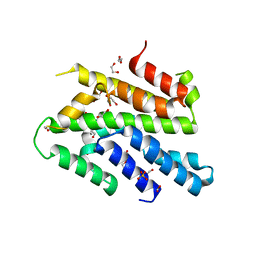 | | Crystal Structure of the Sld3/Treslin domain from yeast Sld3 | | 分子名称: | 1,2-ETHANEDIOL, DNA replication regulator SLD3, SULFATE ION | | 著者 | Itou, H, Araki, H, Shirakihara, Y. | | 登録日 | 2013-09-05 | | 公開日 | 2014-08-20 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of the homology domain of the eukaryotic DNA replication proteins sld3/treslin.
Structure, 22, 2014
|
|
3X37
 
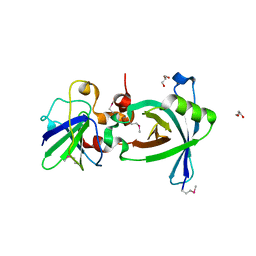 | | Crystal structure of the N-terminal domain of Sld7 in complex with Sld3 | | 分子名称: | GLYCEROL, Mitochondrial morphogenesis protein SLD7, ZYRO0C14696p | | 著者 | Itou, H, Araki, H, Shirakihara, Y. | | 登録日 | 2015-01-16 | | 公開日 | 2015-08-19 | | 最終更新日 | 2017-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | The quaternary structure of the eukaryotic DNA replication proteins Sld7 and Sld3.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3X38
 
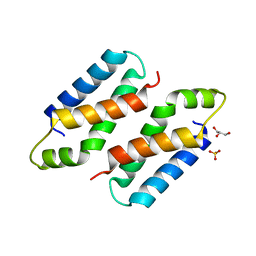 | | Crystal structure of the C-terminal domain of Sld7 | | 分子名称: | GLYCEROL, Mitochondrial morphogenesis protein SLD7, SULFATE ION | | 著者 | Itou, H, Araki, H, Shirakihara, Y. | | 登録日 | 2015-01-16 | | 公開日 | 2015-08-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.801 Å) | | 主引用文献 | The quaternary structure of the eukaryotic DNA replication proteins Sld7 and Sld3.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3AGF
 
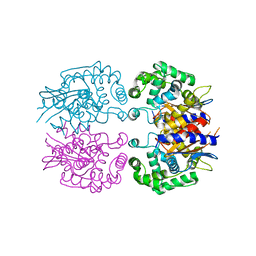 | |
3AGE
 
 | |
