3WR2
 
 | |
4GAD
 
 | |
6XEU
 
 | | CryoEM structure of GIRK2PIP2* - G protein-gated inwardly rectifying potassium channel GIRK2 with PIP2 | | Descriptor: | G protein-activated inward rectifier potassium channel 2, POTASSIUM ION, SODIUM ION, ... | | Authors: | Mathiharan, Y.K, Glaaser, I.W, Skiniotis, G, Slesinger, P.A. | | Deposit date: | 2020-06-13 | | Release date: | 2021-09-01 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into GIRK2 channel modulation by cholesterol and PIP2
Cell Rep, 36, 2021
|
|
4RYT
 
 | |
4XGB
 
 | | Crystal Structure of E112A/H234A Mutant of Stationary Phase Survival Protein (SurE) from Salmonella typhimurium co-crystallized with AMP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5'/3'-nucleotidase SurE, MAGNESIUM ION, ... | | Authors: | Mathiharan, Y.K, Murthy, M.R.N. | | Deposit date: | 2014-12-30 | | Release date: | 2015-09-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Insights into stabilizing interactions in the distorted domain-swapped dimer of Salmonella typhimurium survival protein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XD7
 
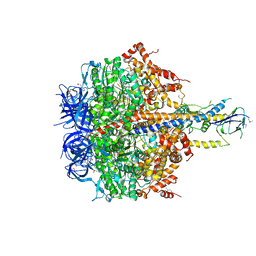 | | Structure of thermophilic F1-ATPase inhibited by epsilon subunit | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase epsilon chain, ATP synthase gamma chain, ... | | Authors: | SHIRAKIHARA, Y, SHIRATORI, A, TANIKAWA, H, NAKASAKO, M, YOSHIDA, M, SUZUKI, T. | | Deposit date: | 2014-12-19 | | Release date: | 2015-08-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structure of a thermophilic F1 -ATPase inhibited by an epsilon-subunit: deeper insight into the epsilon-inhibition mechanism.
Febs J., 282, 2015
|
|
4XGP
 
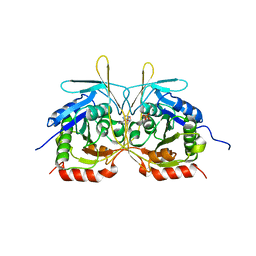 | | Crystal Structure of E112A/H234A Mutant of Stationary Phase Survival Protein (SurE) from Salmonella typhimurium co-crystallized and soaked with AMP. | | Descriptor: | 1,2-ETHANEDIOL, 5'/3'-nucleotidase SurE, ADENINE, ... | | Authors: | Mathiharan, Y.K, Murthy, M.R.N. | | Deposit date: | 2015-01-01 | | Release date: | 2015-09-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into stabilizing interactions in the distorted domain-swapped dimer of Salmonella typhimurium survival protein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XJ7
 
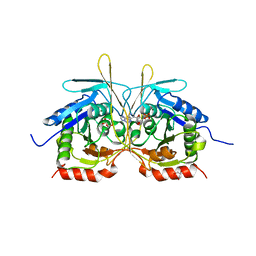 | |
4XER
 
 | | Crystal Structure of C2 form of E112A/H234A Mutant of Stationary Phase Survival Protein (SurE) from Salmonella typhimurium | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5'/3'-nucleotidase SurE, ACETATE ION, ... | | Authors: | Mathiharan, Y.K, Murthy, M.R.N. | | Deposit date: | 2014-12-24 | | Release date: | 2015-09-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Insights into stabilizing interactions in the distorted domain-swapped dimer of Salmonella typhimurium survival protein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XH8
 
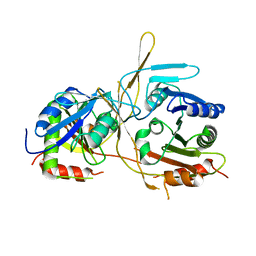 | |
6XEV
 
 | | CryoEM structure of GIRK2-PIP2/CHS - G protein-gated inwardly rectifying potassium channel GIRK2 with modulators cholesteryl hemisuccinate and PIP2 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, G protein-activated inward rectifier potassium channel 2, POTASSIUM ION, ... | | Authors: | Mathiharan, Y.K, Glaaser, I.W, Skiniotis, G, Slesinger, P.A. | | Deposit date: | 2020-06-14 | | Release date: | 2021-09-01 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into GIRK2 channel modulation by cholesterol and PIP2
Cell Rep, 36, 2021
|
|
2RPB
 
 | |
4XEP
 
 | | Crystal Structure of F222 form of E112A/H234A Mutant of Stationary Phase Survival Protein (SurE) from Salmonella typhimurium | | Descriptor: | 1,2-ETHANEDIOL, 5'/3'-nucleotidase SurE, MAGNESIUM ION, ... | | Authors: | Mathiharan, Y.K, Murthy, M.R.N. | | Deposit date: | 2014-12-24 | | Release date: | 2015-09-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Insights into stabilizing interactions in the distorted domain-swapped dimer of Salmonella typhimurium survival protein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4RYU
 
 | |
4G9O
 
 | |
3WMR
 
 | | Crystal structure of VinJ | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, GLYCEROL, Proline iminopeptidase | | Authors: | Shinohara, Y, Miyanaga, A, Kudo, F, Eguchi, T. | | Deposit date: | 2013-11-22 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of the amidohydrolase VinJ shows a unique hydrophobic tunnel for its interaction with polyketide substrates
Febs Lett., 588, 2014
|
|
1PFK
 
 | |
2EXD
 
 | | The solution structure of the C-terminal domain of a nfeD homolog from Pyrococcus horikoshii | | Descriptor: | nfeD short homolog | | Authors: | Kuwahara, Y, Ohno, A, Morii, T, Tochio, H, Shirakawa, M, Hiroaki, H. | | Deposit date: | 2005-11-08 | | Release date: | 2006-12-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the C-terminal domain of NfeD reveals a novel membrane-anchored OB-fold.
Protein Sci., 17, 2008
|
|
8X5U
 
 | | Crystal structure of Thermus thermophilus peptidyl-tRNA hydrolase C-terminal 16 amino acid deletion mutant | | Descriptor: | Peptidyl-tRNA hydrolase, SULFATE ION | | Authors: | Uehara, Y, Matsumoto, A, Nakazawa, T, Fukuta, A, Ando, K, Oka, N, Uchiumi, T, Ito, K. | | Deposit date: | 2023-11-19 | | Release date: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Thermus thermophilus peptidyl-tRNA hydrolase C-terminal 16 amino acid deletion mutant
To Be Published
|
|
8X5T
 
 | | Crystal structure of Thermus thermophilus peptidyl-tRNA hydrolase in complex with adenosine 5'-monophosphate | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Uehara, Y, Matsumoto, A, Nakazawa, T, Fukuta, A, Ando, K, Oka, N, Uchiumi, T, Ito, K. | | Deposit date: | 2023-11-19 | | Release date: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Thermus thermophilus peptidyl-tRNA hydrolase in complex with adenosine 5'-monophosphate
To Be Published
|
|
2DQM
 
 | | Crystal Structure of Aminopeptidase N complexed with bestatin | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, Aminopeptidase N, SULFATE ION, ... | | Authors: | Onohara, Y, Nakajima, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2006-05-29 | | Release date: | 2006-08-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Aminopeptidase N (proteobacteria alanyl aminopeptidase) from Escherichia coli: Crystal structure and conformational change of the methionine 260 residue involved in substrate recognition
J.Biol.Chem., 281, 2006
|
|
1SKY
 
 | | CRYSTAL STRUCTURE OF THE NUCLEOTIDE FREE ALPHA3BETA3 SUB-COMPLEX OF F1-ATPASE FROM THE THERMOPHILIC BACILLUS PS3 | | Descriptor: | F1-ATPASE, SULFATE ION | | Authors: | Shirakihara, Y, Leslie, A.G.W, Abrahams, J.P, Walker, J.E, Ueda, T, Sekimoto, Y, Kambara, M, Saika, K, Kagawa, Y, Yoshida, M. | | Deposit date: | 1997-02-26 | | Release date: | 1998-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The crystal structure of the nucleotide-free alpha 3 beta 3 subcomplex of F1-ATPase from the thermophilic Bacillus PS3 is a symmetric trimer.
Structure, 5, 1997
|
|
8XGK
 
 | | Optimization Efforts for Identification of Novel Highly Potent Keap1-Nrf2 Protein-Protein Interaction Ihhibitors | | Descriptor: | (2~{R},3~{S})-3-[[(2~{S})-2-(4-chlorophenyl)-2-fluoranyl-ethanoyl]amino]-3-[3-(2-cyano-2-methyl-propoxy)-4-methyl-phenyl]-2-methyl-propanoic acid, ACETATE ION, Kelch-like ECH-associated protein 1, ... | | Authors: | Otake, K, Hara, Y, Ubukata, M, Inoue, M, Nagahashi, N, Motoda, D, Ogawa, N, Hantani, Y, Hantani, R, Adachi, T, Nomura, A, Yamaguchi, K, Maekawa, M, Mamada, H, Motomura, T, Sato, M, Harada, K. | | Deposit date: | 2023-12-15 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Optimization Efforts for Identification of Novel Highly Potent Keap1-Nrf2 Protein-Protein Interaction Inhibitors.
J.Med.Chem., 67, 2024
|
|
8XGV
 
 | | Optimization Efforts for Identification of Novel Highly Potent Keap1-Nrf2 Protein-Protein Interaction (PPI) Inhibitors | | Descriptor: | (2~{R},3~{S})-3-[[(2~{S})-2-[4-[(3-ethoxypyridin-2-yl)methyl]phenyl]-2-fluoranyl-ethanoyl]amino]-2-methyl-3-(4-methylphenyl)propanoic acid, ACETATE ION, Kelch-like ECH-associated protein 1, ... | | Authors: | Otake, K, Hara, Y, Ubukata, M, Inoue, M, Nagahashi, N, Motoda, D, Ogawa, N, Hantani, Y, Hantani, R, Adachi, T, Nomura, A, Yamaguchi, K, Maekawa, M, Mamada, H, Motomura, T, Sato, M, Harada, K. | | Deposit date: | 2023-12-15 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Optimization Efforts for Identification of Novel Highly Potent Keap1-Nrf2 Protein-Protein Interaction Inhibitors.
J.Med.Chem., 67, 2024
|
|
3WHO
 
 | | X-ray-Crystallographic Structure of an RNase Po1 Exhibiting Anti-tumor Activity | | Descriptor: | Guanyl-specific ribonuclease Po1 | | Authors: | Kobayashi, H, Katsurtani, T, Hara, Y, Motoyoshi, N, Itagaki, T, Akita, F, Higashiura, A, Yamada, Y, Suzuki, M, Inokuchi, N. | | Deposit date: | 2013-08-30 | | Release date: | 2014-07-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray crystallographic structure of RNase Po1 that exhibits anti-tumor activity.
Biol.Pharm.Bull., 37, 2014
|
|
