5X50
 
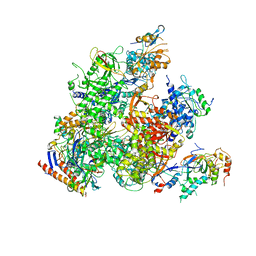 | | RNA Polymerase II from Komagataella Pastoris (Type-2 crystal) | | Descriptor: | DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, RNA polymerase II subunit, ... | | Authors: | Ehara, H, Umehara, T, Sekine, S, Yokoyama, S. | | Deposit date: | 2017-02-14 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4.293 Å) | | Cite: | Crystal structure of RNA polymerase II from Komagataella pastoris
Biochem. Biophys. Res. Commun., 487, 2017
|
|
2CWH
 
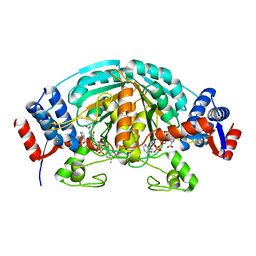 | | Crystal Structure of delta1-piperideine-2-carboxylate reductase from Pseudomonas syringae complexed with NADPH and pyrrole-2-carboxylate | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PYRROLE-2-CARBOXYLATE, delta1-piperideine-2-carboxylate reductase | | Authors: | Goto, M, Muramatsu, H, Mihara, H, Kurihara, T, Esaki, N, Omi, R, Miyahara, I, Hirotsu, K. | | Deposit date: | 2005-06-20 | | Release date: | 2005-10-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of Delta1-piperideine-2-carboxylate/Delta1-pyrroline-2-carboxylate reductase belonging to a new family of NAD(P)H-dependent oxidoreductases: conformational change, substrate recognition, and stereochemistry of the reaction
J.Biol.Chem., 280, 2005
|
|
6T72
 
 | | Structure of the RsaA N-terminal domain bound to LPS | | Descriptor: | 4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose, CALCIUM ION, S-layer protein | | Authors: | von Kuegelgen, A, Bharat, T.A.M. | | Deposit date: | 2019-10-21 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | In Situ Structure of an Intact Lipopolysaccharide-Bound Bacterial Surface Layer.
Cell, 180, 2020
|
|
5WRM
 
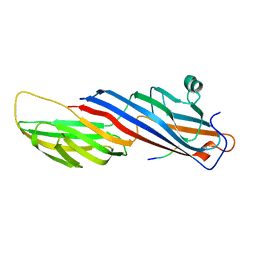 | | Mu2 subunit of the clathrin adaptor complex AP2 in complex with IRS-1 Y658 peptide | | Descriptor: | AP-2 complex subunit mu, Insulin receptor substrate 1 | | Authors: | Yoneyama, Y, Niwa, H, Umehara, T, Yokoyama, S, Hakuno, F, Takahashi, S. | | Deposit date: | 2016-12-02 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | IRS-1 acts as an endocytic regulator of IGF-I receptor to facilitate sustained IGF signaling
Elife, 7, 2018
|
|
1WDW
 
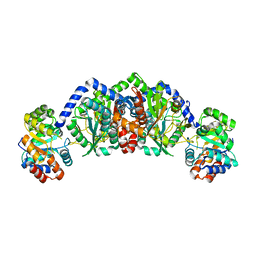 | | Structural basis of mutual activation of the tryptophan synthase a2b2 complex from a hyperthermophile, Pyrococcus furiosus | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Tryptophan synthase alpha chain, Tryptophan synthase beta chain 1 | | Authors: | Lee, S.J, Ogasahara, K, Ma, J, Nishio, K, Ishida, M, Yamagata, Y, Tsukihara, T, Yutani, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-19 | | Release date: | 2005-07-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Conformational Changes in the Tryptophan Synthase from a Hyperthermophile upon alpha(2)beta(2) Complex Formation: Crystal Structure of the Complex
Biochemistry, 44, 2005
|
|
5H34
 
 | | Crystal structure of the C-terminal domain of methionyl-tRNA synthetase (MetRS-C) in Nanoarchaeum equitans | | Descriptor: | Methionine-tRNA ligase | | Authors: | Suzuki, H, Kaneko, A, Yamamoto, T, Nambo, M, Umehara, T, Yoshida, H, Park, S.Y, Tamura, K. | | Deposit date: | 2016-10-20 | | Release date: | 2017-06-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.748 Å) | | Cite: | Binding Properties of Split tRNA to the C-terminal Domain of Methionyl-tRNA Synthetase of Nanoarchaeum equitans.
J. Mol. Evol., 84, 2017
|
|
7EIC
 
 | |
7EID
 
 | |
7EIE
 
 | | Crystal structure of YEATS2 YEATS domain | | Descriptor: | GLYCEROL, YEATS domain-containing protein 2 | | Authors: | Kikuchi, M, Umehara, T. | | Deposit date: | 2021-03-30 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Elucidation of binding preferences of YEATS domains to site-specific acetylated nucleosome core particles.
J.Biol.Chem., 298, 2022
|
|
2RR6
 
 | | Solution structure of the leucine rich repeat of human acidic leucine-rich nuclear phosphoprotein 32 family member B | | Descriptor: | Acidic leucine-rich nuclear phosphoprotein 32 family member B | | Authors: | Tochio, N, Umehara, T, Tsuda, K, Koshiba, S, Harada, T, Watanabe, S, Tanaka, A, Kigawa, T, Yokoyama, S. | | Deposit date: | 2010-05-25 | | Release date: | 2010-06-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of histone chaperone ANP32B: interaction with core histones H3-H4 through its acidic concave domain.
J.Mol.Biol., 401, 2010
|
|
4BH5
 
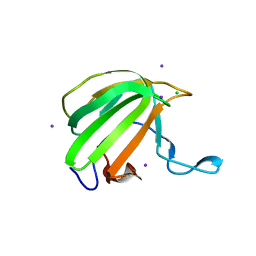 | | LytM domain of EnvC, an activator of cell wall amidases in Escherichia coli | | Descriptor: | CHLORIDE ION, GLYCEROL, IODIDE ION, ... | | Authors: | Morlot, C, Peters, N.T, Yang, D.C, Uehara, T, Vernet, T, Bernhardt, T.G. | | Deposit date: | 2013-03-29 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structure-Function Analysis of the Lytm Domain of Envc, an Activator of Cell Wall Remodeling at the Escherichia Coli Division Site.
Mol.Microbiol., 89, 2013
|
|
2YRP
 
 | | Solution structure of the TIG domain from Human Nuclear factor of activated T-cells, cytoplasmic 4 | | Descriptor: | Nuclear factor of activated T-cells, cytoplasmic 4 | | Authors: | Tomizawa, T, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-02 | | Release date: | 2008-02-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the TIG domain from Human Nuclear factor of activated T-cells, cytoplasmic 4
To be Published
|
|
1NC8
 
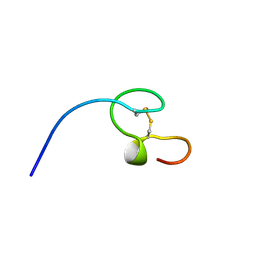 | | HIGH-RESOLUTION SOLUTION NMR STRUCTURE OF THE MINIMAL ACTIVE DOMAIN OF THE HUMAN IMMUNODEFICIENCY VIRUS TYPE-2 NUCLEOCAPSID PROTEIN, 15 STRUCTURES | | Descriptor: | NUCLEOCAPSID PROTEIN, ZINC ION | | Authors: | Kodera, Y, Sato, K, Tsukahara, T, Komatsu, H, Maeda, T, Kohno, T. | | Deposit date: | 1998-05-14 | | Release date: | 1999-05-25 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution NMR structure of the minimal active domain of the human immunodeficiency virus type-2 nucleocapsid protein.
Biochemistry, 37, 1998
|
|
5H6R
 
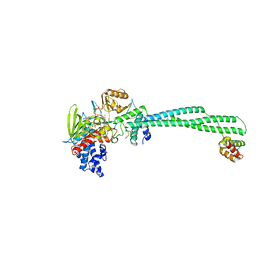 | | Crystal structure of LSD1-CoREST in complex with peptide 13 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1A, ... | | Authors: | Kkuchi, M, Amano, Y, Sato, S, Yokoyama, S, Umezawa, N, Higuchi, T, Umehara, T. | | Deposit date: | 2016-11-14 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Development and crystallographic evaluation of histone H3 peptide with N-terminal serine substitution as a potent inhibitor of lysine-specific demethylase 1.
Bioorg. Med. Chem., 25, 2017
|
|
5H6Q
 
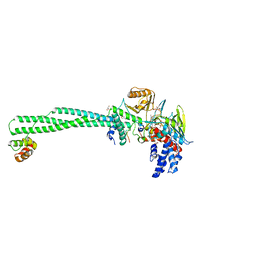 | | Crystal structure of LSD1-CoREST in complex with peptide 11 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1A, ... | | Authors: | Kikuchi, M, Amano, Y, Sato, S, Yokoyama, S, Umezawa, N, Higuchi, T, Umehara, T. | | Deposit date: | 2016-11-14 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Development and crystallographic evaluation of histone H3 peptide with N-terminal serine substitution as a potent inhibitor of lysine-specific demethylase 1.
Bioorg. Med. Chem., 25, 2017
|
|
1V6C
 
 | | Crystal Structure of Psychrophilic Subtilisin-like Protease Apa1 from Antarctic Psychrotroph Pseudoalteromonas sp. AS-11 | | Descriptor: | CALCIUM ION, SULFATE ION, alkaline serine protease, ... | | Authors: | Dong, D, Ihara, T, Motoshima, H, Watanabe, K. | | Deposit date: | 2003-11-28 | | Release date: | 2004-12-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Psychrophilic Subtilisin-like Protease Apa1 from Antarctic Psychrotroph Pseudoalteromonas sp. AS-11
To be Published
|
|
2YT2
 
 | | Solution structure of the chimera of the PTB domain of SNT-2 and 19-residue peptide (aa 1571-1589) of hALK | | Descriptor: | Fibroblast growth factor receptor substrate 3 and ALK tyrosine kinase receptor | | Authors: | Li, H, Koshiba, S, Tomizawa, T, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of nucleophosmin-anaplastic lymphoma kinase oncoprotein by the phosphotyrosine binding domain of Suc1-associated neurotrophic factor-induced tyrosine-phosphorylated target-2
J.Struct.Funct.Genom., 11, 2010
|
|
1CYC
 
 | | THE CRYSTAL STRUCTURE OF BONITO (KATSUO) FERROCYTOCHROME C AT 2.3 ANGSTROMS RESOLUTION. II. STRUCTURE AND FUNCTION | | Descriptor: | FERROCYTOCHROME C, HEME C | | Authors: | Tanaka, N, Yamane, T, Tsukihara, T, Ashida, T, Kakudo, M. | | Deposit date: | 1976-08-01 | | Release date: | 1976-10-06 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of bonito (katsuo) ferrocytochrome c at 2.3 A resolution. II. Structure and function.
J.Biochem.(Tokyo), 77, 1975
|
|
2YSH
 
 | | Solution structure of the WW domain from the human growth-arrest-specific protein 7, GAS-7 | | Descriptor: | Growth-arrest-specific protein 7 | | Authors: | Ohnishi, S, Tochio, N, Sato, M, Koshiba, S, Harada, T, Watanabe, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the WW domain from the human growth-arrest-specific protein 7, GAS-7
To be Published
|
|
2YSX
 
 | | Solution structure of the human SHIP SH2 domain | | Descriptor: | Signaling inositol polyphosphate phosphatase SHIP II | | Authors: | Kasai, T, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-04 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the human SHIP SH2 domain
To be Published
|
|
2YS9
 
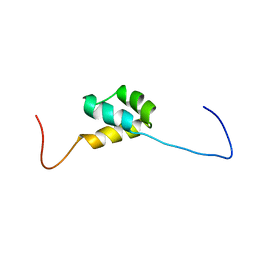 | | structure of the third Homeodomain from the human homeobox and leucine zipper protein, Homez | | Descriptor: | Homeobox and leucine zipper protein Homez | | Authors: | Ohnishi, S, Tomizawa, T, Koshiba, S, Harada, T, Watanabe, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | structure of the third Homeodomain from the human homeobox and leucine zipper protein, Homez
To be Published
|
|
2YSO
 
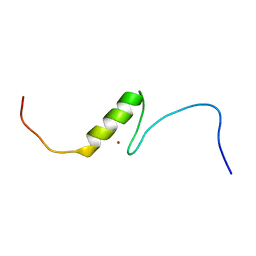 | | Solution structure of the C2H2 type zinc finger (region 656-688) of human Zinc finger protein 95 homolog | | Descriptor: | ZINC ION, Zinc finger protein 95 homolog | | Authors: | Takahashi, M, Kuwasako, K, Tsuda, K, Tanabe, W, Harada, T, Watanabe, S, Tochio, N, Muto, Y, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C2H2 type zinc finger (region 656-688) of human Zinc finger protein 95 homolog
To be Published
|
|
2YTV
 
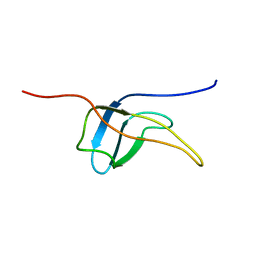 | | Solution structure of the fifth cold-shock domain of the human KIAA0885 protein (unr protein) | | Descriptor: | Cold shock domain-containing protein E1 | | Authors: | Goroncy, A.K, Tochio, N, Tomizawa, T, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structures of the five constituent cold-shock domains (CSD) of the human UNR (upstream of N-ras) protein.
J.Struct.Funct.Genom., 11, 2010
|
|
2YUQ
 
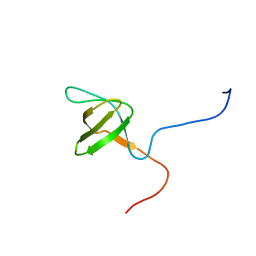 | | Solution structure of the SH3 domain of human Tyrosine-protein kinase ITK/TSK | | Descriptor: | Tyrosine-protein kinase ITK/TSK | | Authors: | Abe, H, Miyamoto, K, Tochio, N, Sato, M, Koshiba, S, Harada, T, Watanabe, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-06 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain of human Tyrosine-protein kinase ITK/TSK
To be Published
|
|
2YRQ
 
 | | Solution structure of the tandem HMG box domain from Human High mobility group protein B1 | | Descriptor: | High mobility group protein B1 | | Authors: | Tomizawa, T, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-02 | | Release date: | 2008-02-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the tandem HMG box domain from Human High mobility group protein B1
To be Published
|
|
