8SL1
 
 | | Cryo-EM structure of PAPP-A2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Judge, R.A, Stoll, V.S, Eaton, D, Hao, Q, Bratkowski, M.A. | | Deposit date: | 2023-04-20 | | Release date: | 2023-11-08 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Cryo-EM structure of human PAPP-A2 and mechanism of substrate recognition.
Commun Chem, 6, 2023
|
|
8GCR
 
 | | HPV16 E6-E6AP-p53 complex | | Descriptor: | Cellular tumor antigen p53, Maltose/maltodextrin-binding periplasmic protein,Protein E6, Ubiquitin-protein ligase E3A, ... | | Authors: | Bratkowski, M.A, Wang, J.C.K, Hao, Q, Nile, A.H. | | Deposit date: | 2023-03-02 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structure of the p53 degradation complex from HPV16.
Nat Commun, 15, 2024
|
|
7CQT
 
 | | Crystal structure of Brassica juncea HMG-CoA synthase 1 mutant - S359A in complex with acetyl-CoA | | Descriptor: | 3-hydroxy-3-methylglutaryl coenzyme A synthase, ACETYL COENZYME *A, ADENOSINE-3'-5'-DIPHOSPHATE | | Authors: | Liao, P, Hu, M, Kong, G.K.W, Hao, Q, Chye, M.L. | | Deposit date: | 2020-08-11 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Overexpression and Inhibition of 3-Hydroxy-3-Methylglutaryl-CoA Synthase Affect Central Metabolic Pathways in Tobacco.
Plant Cell.Physiol., 62, 2021
|
|
8Y1R
 
 | | in situ room temperature Laue crystallography | | Descriptor: | Lysozyme C | | Authors: | Wang, Z.J, Wang, S.S, Pan, Q.Y, Yu, L, Su, Z.H, Yang, T.Y, Wang, Y.Z, Zhang, W.Z, Hao, Q, Gao, X.Y. | | Deposit date: | 2024-01-25 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | BL03HB: Laue crystallography beamline at SSRF
To Be Published
|
|
8W6K
 
 | | in situ room temperature Laue crystallography | | Descriptor: | Lysozyme C | | Authors: | Wang, Z.J, Wang, S.S, Pan, Q.Y, Yu, L, Su, Z.H, Yang, T.Y, Wang, Y.Z, Zhang, W.Z, Hao, Q, Gao, X.Y. | | Deposit date: | 2023-08-29 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | BL03HB: Laue crystallography beamline at SSRF
To Be Published
|
|
8HLY
 
 | |
8HLW
 
 | | Crystal structure of SIRT3 in complex with H4K16la peptide | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, Histone H4 residues 20-27, NAD-dependent protein deacetylase sirtuin-3, ... | | Authors: | Zhuming, F, Hao, Q. | | Deposit date: | 2022-12-01 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of SIRT3 as an eraser of H4K16la.
Iscience, 26, 2023
|
|
5WTD
 
 | | Structure of human serum transferrin bound ruthenium at N-lobe | | Descriptor: | FE (III) ION, MALONATE ION, RUTHENIUM ION, ... | | Authors: | Sun, H, Wang, M, Lai, T.P, Zhang, H, Hao, Q. | | Deposit date: | 2016-12-11 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Binding of ruthenium and osmium at non‐iron sites of transferrin accounts for their iron-independent cellular uptake.
J.Inorg.Biochem., 234, 2022
|
|
5WYI
 
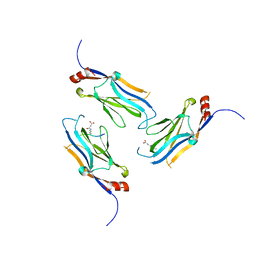 | | The Yaf9 YEATS domain Recognizing H3K122suc Peptide | | Descriptor: | (2S)-2-azanyl-6-[(4-hydroxy-4-oxo-butanoyl)amino]hexanoic acid, ILE-MET-PRO-LYS-ASP-ILE-GLN-LEU, SUCCINIC ACID, ... | | Authors: | Wang, Y, Hao, Q. | | Deposit date: | 2017-01-13 | | Release date: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Yaf9 YEATS domain Recognizing H3K122suc Peptide
To Be Published
|
|
5Y8V
 
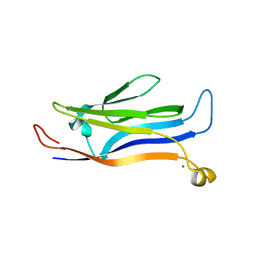 | | Crystal structure of GAS41 | | Descriptor: | MAGNESIUM ION, YEATS domain-containing protein 4 | | Authors: | Wang, Y, Hao, Q. | | Deposit date: | 2017-08-21 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Identification of the YEATS domain of GAS41 as a pH-dependent reader of histone succinylation
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5ZGU
 
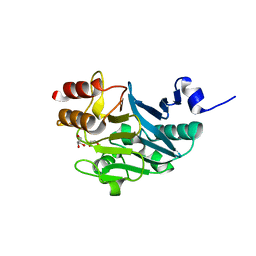 | |
5ZJC
 
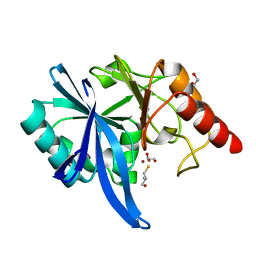 | |
5ZGR
 
 | | Crystal structure of NDM-1 at pH7.3 (HEPES) in complex with hydrolyzed ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, GLYCEROL, Metallo-beta-lactamase type 2, ... | | Authors: | Zhang, H, Hao, Q. | | Deposit date: | 2018-03-10 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Active-Site Conformational Fluctuations Promote the Enzymatic Activity of NDM-1.
Antimicrob. Agents Chemother., 62, 2018
|
|
5ZGV
 
 | |
5ZJ7
 
 | |
5ZGY
 
 | |
5ZIO
 
 | | Crystal structure of NDM-1 in complex with L-captopril | | Descriptor: | L-CAPTOPRIL, Metallo-beta-lactamase type 2, ZINC ION | | Authors: | Zhang, H, Hao, Q. | | Deposit date: | 2018-03-16 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Structure-guided optimization of D-captopril for discovery of potent NDM-1 inhibitors
Bioorg.Med.Chem., 29, 2020
|
|
5ZGW
 
 | |
5ZJ2
 
 | | Crystal structure of NDM-1 in complex with D-captopril | | Descriptor: | 1-(3-MERCAPTO-2-METHYL-PROPIONYL)-PYRROLIDINE-2-CARBOXYLIC ACID, Metallo-beta-lactamase type 2, ZINC ION | | Authors: | Zhang, H, Hao, Q. | | Deposit date: | 2018-03-18 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure-guided optimization of D-captopril for discovery of potent NDM-1 inhibitors
Bioorg.Med.Chem., 29, 2020
|
|
5ZGT
 
 | |
5ZJ1
 
 | |
5ZJ8
 
 | |
5ZH1
 
 | |
5ZGQ
 
 | | Crystal structure of NDM-1 at pH7.5 (Tris-HCl, (NH4)2SO4) in complex with hydrolyzed ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, HYDROXIDE ION, Metallo-beta-lactamase type 2, ... | | Authors: | Zhang, H, Hao, Q. | | Deposit date: | 2018-03-10 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Active-Site Conformational Fluctuations Promote the Enzymatic Activity of NDM-1.
Antimicrob. Agents Chemother., 62, 2018
|
|
5ZGZ
 
 | |
