7DPA
 
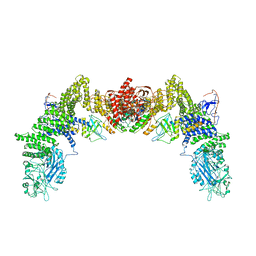 | | Cryo-EM structure of the human ELMO1-DOCK5-Rac1 complex | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Kukimoto-Niino, M, Katsura, K, Kaushik, R, Ehara, H, Yokoyama, T, Uchikubo-Kamo, T, Mishima-Tsumagari, C, Yonemochi, M, Ikeda, M, Hanada, K, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2020-12-18 | | Release date: | 2021-08-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of the human ELMO1-DOCK5-Rac1 complex.
Sci Adv, 7, 2021
|
|
7DVC
 
 | | Crystal structure of the computationally designed reDPBB_sym1 protein | | Descriptor: | ACETATE ION, CHLORIDE ION, reDPBB_sym1 protein | | Authors: | Yagi, S, Tagami, S, Padhi, A.K, Zhang, K.Y.J. | | Deposit date: | 2021-01-13 | | Release date: | 2021-09-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | Seven Amino Acid Types Suffice to Create the Core Fold of RNA Polymerase.
J.Am.Chem.Soc., 143, 2021
|
|
7W18
 
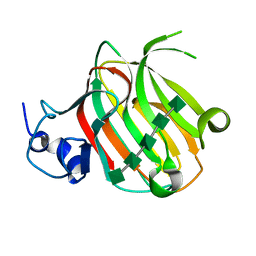 | | Complex structure of alginate lyase PyAly with M5 | | Descriptor: | Alginate lyase, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Liu, W.Z, Lyu, Q.Q, Zhang, K.K. | | Deposit date: | 2021-11-19 | | Release date: | 2022-08-17 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Determination of oligosaccharide product distributions of PL7 alginate lyases by their structural elements.
Commun Biol, 5, 2022
|
|
7W12
 
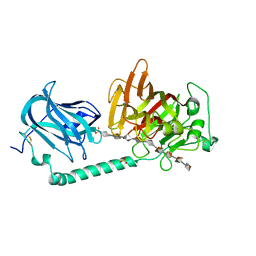 | | Complex structure of alginate lyase AlyB-OU02 with G9 | | Descriptor: | Alginate lyase, SULFATE ION, alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid | | Authors: | Liu, W.Z, Lyu, Q.Q, Zhang, K.K. | | Deposit date: | 2021-11-19 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Determination of oligosaccharide product distributions of PL7 alginate lyases by their structural elements.
Commun Biol, 5, 2022
|
|
7W16
 
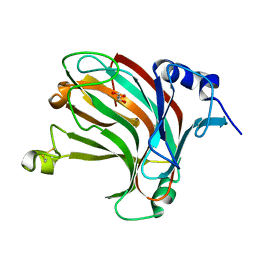 | | Complex structure of alginate lyase AlyV with M8 | | Descriptor: | GLYCEROL, alginate lyase, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Liu, W.Z, Lyu, Q.Q, Li, Z.J, Zhang, K.K. | | Deposit date: | 2021-11-19 | | Release date: | 2022-08-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Determination of oligosaccharide product distributions of PL7 alginate lyases by their structural elements.
Commun Biol, 5, 2022
|
|
7W13
 
 | | Complex structure of alginate lyase PyAly with M8 | | Descriptor: | Alginate lyase, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Liu, W.Z, Lyu, Q.Q, Li, Z.J, Zhang, K.K. | | Deposit date: | 2021-11-19 | | Release date: | 2022-08-17 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Determination of oligosaccharide product distributions of PL7 alginate lyases by their structural elements.
Commun Biol, 5, 2022
|
|
4FK3
 
 | | B-Raf Kinase V600E Oncogenic Mutant in Complex with PLX3203 | | Descriptor: | N-{2,4-difluoro-3-[(5-pyridin-3-yl-1H-pyrrolo[2,3-b]pyridin-3-yl)carbonyl]phenyl}ethanesulfonamide, Serine/threonine-protein kinase B-raf | | Authors: | Zhang, Y, Wang, W, Zhang, K.Y.J. | | Deposit date: | 2012-06-12 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
1T4I
 
 | | Crystal Structure of a DNA Decamer Containing a Thymine-dimer | | Descriptor: | 5'-D(*CP*GP*AP*AP*TP*TP*AP*AP*GP*C)-3', 5'-D(*GP*CP*TP*TP*AP*AP*TP*TP*CP*G)-3' | | Authors: | Park, H, Zhang, K, Ren, Y, Nadji, S, Sinha, N, Taylor, J.S, Kang, C. | | Deposit date: | 2004-04-29 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a DNA Decamer Containing a cis-syn Thymine-dimer
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
7XML
 
 | | Cryo-EM structure of PEIP-Bs_enolase complex | | Descriptor: | Enolase, MAGNESIUM ION, Putative gene 60 protein | | Authors: | Li, S, Zhang, K. | | Deposit date: | 2022-04-26 | | Release date: | 2022-07-27 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Bacteriophage protein PEIP is a potent Bacillus subtilis enolase inhibitor.
Cell Rep, 40, 2022
|
|
1HOX
 
 | | CRYSTAL STRUCTURE OF RABBIT PHOSPHOGLUCOSE ISOMERASE COMPLEXED WITH FRUCTOSE-6-PHOSPHATE | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, PHOSPHOGLUCOSE ISOMERASE | | Authors: | Jeffrey, C.J, Lee, J.H, Chang, K.Z, Patel, V. | | Deposit date: | 2000-12-11 | | Release date: | 2001-07-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of rabbit phosphoglucose isomerase complexed with its substrate D-fructose 6-phosphate.
Biochemistry, 40, 2001
|
|
7EZ0
 
 | | Apo L-21 ScaI Tetrahymena ribozyme | | Descriptor: | Apo L-21 ScaI Tetrahymena ribozyme, MAGNESIUM ION | | Authors: | Su, Z, Zhang, K, Kappel, K, Luo, B, Das, R, Chiu, W. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-25 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Cryo-EM structures of full-length Tetrahymena ribozyme at 3.1 angstrom resolution.
Nature, 596, 2021
|
|
7EZ2
 
 | | Holo L-16 ScaI Tetrahymena ribozyme | | Descriptor: | Holo L-16 ScaI Tetrahymena ribozyme, Holo L-16 ScaI Tetrahymena ribozyme S1, Holo L-16 ScaI Tetrahymena ribozyme S2, ... | | Authors: | Su, Z, Zhang, K, Kappel, K, Luo, B, Das, R, Chiu, W. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-25 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Cryo-EM structures of full-length Tetrahymena ribozyme at 3.1 angstrom resolution.
Nature, 596, 2021
|
|
2ESF
 
 | | Identification of a Novel Non-Catalytic Bicarbonate Binding Site in Eubacterial beta-Carbonic Anhydrase | | Descriptor: | BICARBONATE ION, Carbonic anhydrase 2, ZINC ION | | Authors: | Cronk, J.D, Rowlett, R.S, Zhang, K.Y.J, Tu, C, Endrizzi, J.A, Gareiss, P.C. | | Deposit date: | 2005-10-26 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Identification of a Novel Noncatalytic Bicarbonate Binding Site in Eubacterial beta-Carbonic Anhydrase.
Biochemistry, 45, 2006
|
|
5HSI
 
 | | Crystal structure of tyrosine decarboxylase at 1.73 Angstroms resolution | | Descriptor: | MAGNESIUM ION, Putative decarboxylase | | Authors: | Ni, Y, Zhou, J, Zhu, H, Zhang, K. | | Deposit date: | 2016-01-25 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.732 Å) | | Cite: | Crystal structure of tyrosine decarboxylase and identification of key residues involved in conformational swing and substrate binding
Sci Rep, 6, 2016
|
|
5HSJ
 
 | | Structure of tyrosine decarboxylase complex with PLP at 1.9 Angstroms resolution | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Putative decarboxylase | | Authors: | Ni, Y, Zhou, J, Zhu, H, Zhang, K. | | Deposit date: | 2016-01-25 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of tyrosine decarboxylase and identification of key residues involved in conformational swing and substrate binding
Sci Rep, 6, 2016
|
|
1N4E
 
 | | Crystal Structure of a DNA Decamer Containing a Thymine-dimer | | Descriptor: | 5'-D(*CP*GP*AP*AP*TP*TP*AP*AP*GP*C)-3', 5'-D(*GP*CP*TP*TP*AP*AP*TP*TP*CP*G)-3' | | Authors: | Park, H, Zhang, K, Ren, Y, Nadji, S, Sinha, N, Taylor, J.-S, Kang, C. | | Deposit date: | 2002-10-30 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a DNA decamer containing a cis-syn thymine dimer.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
4HND
 
 | | Crystal structure of the catalytic domain of Selenomethionine substituted human PI4KIIalpha in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Phosphatidylinositol 4-kinase type 2-alpha | | Authors: | Zhou, Q, Zhai, Y, Zhang, K, Chen, C, Sun, F. | | Deposit date: | 2012-10-19 | | Release date: | 2014-04-09 | | Last modified: | 2016-12-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Molecular insights into the membrane-associated phosphatidylinositol 4-kinase II alpha.
Nat Commun, 5, 2014
|
|
7DVH
 
 | |
7DVF
 
 | | Crystal structure of the computationally designed reDPBB_sym2 protein | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, reDPBB_sym2 protein | | Authors: | Yagi, S, Tagami, S, Padhi, A.K, Zhang, K.Y.J. | | Deposit date: | 2021-01-13 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.209 Å) | | Cite: | Seven Amino Acid Types Suffice to Create the Core Fold of RNA Polymerase.
J.Am.Chem.Soc., 143, 2021
|
|
4HNE
 
 | | Crystal structure of the catalytic domain of human type II alpha Phosphatidylinositol 4-kinase (PI4KIIalpha) in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Phosphatidylinositol 4-kinase type 2-alpha | | Authors: | Zhou, Q, Zhai, Y, Zhang, K, Chen, C, Sun, F. | | Deposit date: | 2012-10-19 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Molecular insights into the membrane-associated phosphatidylinositol 4-kinase II alpha.
Nat Commun, 5, 2014
|
|
1K51
 
 | | A G55A Mutation Induces 3D Domain Swapping in the B1 Domain of Protein L from Peptostreptococcus magnus | | Descriptor: | Protein L, ZINC ION | | Authors: | O'Neill, J.W, Kim, D.E, Johnsen, K, Baker, D, Zhang, K.Y.J. | | Deposit date: | 2001-10-09 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Single-site mutations induce 3D domain swapping in the B1 domain of protein L from Peptostreptococcus magnus.
Structure, 9, 2001
|
|
1K52
 
 | | Monomeric Protein L B1 Domain with a K54G mutation | | Descriptor: | Protein L, ZINC ION | | Authors: | O'Neill, J.W, Kim, D.E, Johnsen, K, Baker, D, Zhang, K.Y.J. | | Deposit date: | 2001-10-09 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Single-site mutations induce 3D domain swapping in the B1 domain of protein L from Peptostreptococcus magnus.
Structure, 9, 2001
|
|
1K50
 
 | | A V49A Mutation Induces 3D Domain Swapping in the B1 Domain of Protein L from Peptostreptococcus magnus | | Descriptor: | Protein L | | Authors: | O'Neill, J.W, Kim, D.E, Johnsen, K, Baker, D, Zhang, K.Y.J. | | Deposit date: | 2001-10-09 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Single-site mutations induce 3D domain swapping in the B1 domain of protein L from Peptostreptococcus magnus.
Structure, 9, 2001
|
|
1K53
 
 | | Monomeric Protein L B1 Domain with a G15A Mutation | | Descriptor: | Protein L, ZINC ION | | Authors: | O'Neill, J.W, Kim, D.E, Johnsen, K, Baker, D, Zhang, K.Y.J. | | Deposit date: | 2001-10-09 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Single-site mutations induce 3D domain swapping in the B1 domain of protein L from Peptostreptococcus magnus.
Structure, 9, 2001
|
|
1Y2E
 
 | | Catalytic Domain Of Human Phosphodiesterase 4D In Complex With 1-(4-amino-phenyl)-3,5-dimethyl-1H-pyrazole-4-carboxylic acid ethyl ester | | Descriptor: | 1,2-ETHANEDIOL, 1-(4-AMINOPHENYL)-3,5-DIMETHYL-1H-PYRAZOLE-4-CARBOXYLIC ACID ETHYL ESTER, MAGNESIUM ION, ... | | Authors: | Card, G.L, Blasdel, L, England, B.P, Zhang, C, Suzuki, Y, Gillette, S, Fong, D, Ibrahim, P.N, Artis, D.R, Bollag, G, Milburn, M.V, Kim, S.-H, Schlessinger, J, Zhang, K.Y.J. | | Deposit date: | 2004-11-22 | | Release date: | 2005-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A family of phosphodiesterase inhibitors discovered by cocrystallography and scaffold-based drug design
Nat.Biotechnol., 23, 2005
|
|
