7V5K
 
 | | MERS S ectodomain trimer in complex with neutralizing antibody 0722 (state 1) | | Descriptor: | 0722 H, 0722 L, Spike glycoprotein | | Authors: | Wang, X, Zhao, J, Wang, Z, Zeng, J, Zhang, S, Wang, Y. | | Deposit date: | 2021-08-17 | | Release date: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | MERS S ectodomain trimer in complex with neutralizing antibody 0722 (state 1)
to be published
|
|
3IKH
 
 | |
8R8N
 
 | |
7V5J
 
 | | MERS S ectodomain trimer in complex with neutralizing antibody 0722(state 2) | | Descriptor: | 0722 H, 0722 L, Spike glycoprotein | | Authors: | Wang, X, Zhao, J, Wang, Z, Zeng, J, Zhang, S, Wang, Y. | | Deposit date: | 2021-08-17 | | Release date: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | MERS S ectodomain trimer in complex with neutralizing antibody 0722(state 2)
to be published
|
|
7V6N
 
 | | MERS S ectodomain trimer in complex with neutralizing antibody 111 state1 | | Descriptor: | 111 H, 111 L, Spike glycoprotein | | Authors: | Wang, X, Zhao, J, Wang, Z, Zeng, J, Zhang, S, Wang, Y. | | Deposit date: | 2021-08-20 | | Release date: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | MERS S ectodomain trimer in complex with neutralizing antibody 111 state1
to be published
|
|
7V6O
 
 | | MERS S ectodomain trimer in complex with neutralizing antibody 111 (state 2) | | Descriptor: | 111 H, 111 L, Spike glycoprotein | | Authors: | Wang, X, Zhao, J, Wang, Z, Zeng, J, Zhang, S, Wang, Y. | | Deposit date: | 2021-08-20 | | Release date: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (4.56 Å) | | Cite: | MERS S ectodomain trimer in complex with neutralizing antibody 111 (state 2)
to be published
|
|
3H75
 
 | | Crystal Structure of a Periplasmic Sugar-binding protein from the Pseudomonas fluorescens | | Descriptor: | GLYCEROL, Periplasmic sugar-binding domain protein, SULFATE ION | | Authors: | Kumaran, D, Mahmood, A, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-24 | | Release date: | 2009-05-12 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of a Periplasmic Sugar-binding protein from the Pseudomonas fluorescens
To be Published
|
|
3HEB
 
 | |
4UCG
 
 | | NmeDAH7PS R126S variant | | Descriptor: | DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, PHOSPHO-2-DEHYDRO-3-DEOXYHEPTONATE ALDOLASE, ... | | Authors: | Cross, P.J, Heyes, L.C, Zhang, S, Nazmi, A.R, Parker, E.J. | | Deposit date: | 2014-12-03 | | Release date: | 2016-01-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Functional Unit of Neisseria Meningitidis 3-Deoxy-D-Arabino-Heptulosonate 7-Phosphate Synthase is Dimeric.
Plos One, 11, 2016
|
|
3ID9
 
 | |
3IE7
 
 | |
3IPI
 
 | | Crystal Structure of a Geranyltranstransferase from the Methanosarcina mazei | | Descriptor: | Geranyltranstransferase, MALONIC ACID | | Authors: | Kumaran, D, Mohammed, M.B, Brown, A, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-17 | | Release date: | 2009-09-08 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of a Geranyltranstransferase from the Methanosarcina mazei
To be Published
|
|
3IBQ
 
 | | Crystal structure of pyridoxal kinase from Lactobacillus plantarum in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Pyridoxal kinase | | Authors: | Bagaria, A, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-16 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of pyridoxal kinase from Lactobacillus plantarum in complex with ATP
To be Published
|
|
3IN1
 
 | |
3ILH
 
 | |
3HZ4
 
 | |
3ILV
 
 | |
3F6A
 
 | |
3FFZ
 
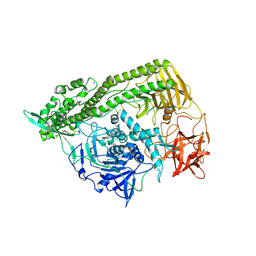 | | Domain organization in Clostridium butulinum neurotoxin type E is unique: Its implication in faster translocation | | Descriptor: | ACETATE ION, Botulinum neurotoxin type E, SODIUM ION, ... | | Authors: | Kumaran, D, Eswaramoorthy, S, Swaminathan, S. | | Deposit date: | 2008-12-04 | | Release date: | 2008-12-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Domain organization in Clostridium botulinum neurotoxin type E is unique: its implication in faster translocation.
J.Mol.Biol., 386, 2009
|
|
3G7S
 
 | |
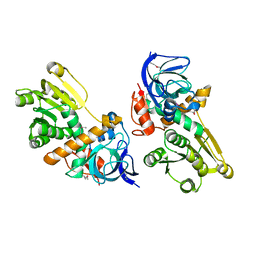
3GAZ
 
 | |
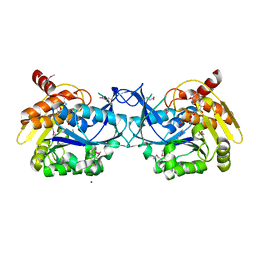
3IQ0
 
 | |
8ILM
 
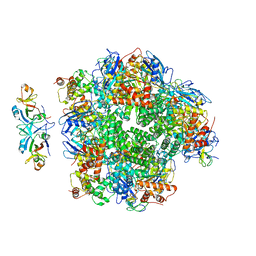 | | The cryo-EM structure of eight Rubisco large subunits (RbcL), two Arabidopsis thaliana Rubisco accumulation factors 1 (AtRaf1), and seven Arabidopsis thaliana Bundle Sheath Defective 2 (AtBSD2) | | Descriptor: | Protein BUNDLE SHEATH DEFECTIVE 2, chloroplastic, Ribulose bisphosphate carboxylase large chain, ... | | Authors: | Wang, R, Song, H, Zhang, W, Wang, N, Zhang, S, Shao, R. | | Deposit date: | 2023-03-03 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the functions of Raf1 and Bsd2 in hexadecameric Rubisco assembly.
Mol Plant, 16, 2023
|
|
7T91
 
 | |
6QP1
 
 | | Crystal structure of the PLP-bound C-S lyase in the external aldimine form from Staphylococcus hominis complexed with an inhibitor, L-cycloserine. | | Descriptor: | Aminotransferase, [5-hydroxy-6-methyl-4-({[(4E)-3-oxo-1,2-oxazolidin-4-ylidene]amino}methyl)pyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Herman, R, Rudden, M, Wilkinson, A.J, Hanai, S, Thomas, G.H. | | Deposit date: | 2019-02-13 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | The molecular basis of thioalcohol production in human body odour.
Sci Rep, 10, 2020
|
|
