1FEX
 
 | |
8GOD
 
 | | Co-crystal structure of Human Protein-arginine deiminase type-4 (PAD4) with small molecule inhibitor JBI-589 | | Descriptor: | Protein-arginine deiminase type-4, [(3~{R})-3-azanylpiperidin-1-yl]-[2-[1-[(4-fluorophenyl)methyl]indol-2-yl]-3-methyl-imidazo[1,2-a]pyridin-7-yl]methanone | | Authors: | Swaminathan, S, Birudukota, S, Vaithilingam, K, Kandan, S, Asaithambi, K, Kathiresan, N, Gosu, R, Rajagopal, S, Sadhu, N. | | Deposit date: | 2022-08-24 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Alleviation of arthritis through prevention of neutrophil extracellular traps by an orally available inhibitor of protein arginine deiminase 4.
Sci Rep, 13, 2023
|
|
3SPC
 
 | | Inward rectifier potassium channel Kir2.2 in complex with dioctanoylglycerol pyrophosphate (DGPP) | | Descriptor: | (2R)-3-{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}propane-1,2-diyl dioctanoate, Inward-rectifier K+ channel Kir2.2, POTASSIUM ION | | Authors: | Hansen, S.B, Tao, X, MacKinnon, R. | | Deposit date: | 2011-07-01 | | Release date: | 2011-08-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.454 Å) | | Cite: | Structural basis of PIP(2) activation of the classical inward rectifier K(+) channel Kir2.2.
Nature, 477, 2011
|
|
3SPG
 
 | | Inward rectifier potassium channel Kir2.2 R186A mutant in complex with PIP2 | | Descriptor: | Inward-rectifier K+ channel Kir2.2, POTASSIUM ION, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Hansen, S.B, Tao, X, MacKinnon, R. | | Deposit date: | 2011-07-01 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.613 Å) | | Cite: | Structural basis of PIP(2) activation of the classical inward rectifier K(+) channel Kir2.2.
Nature, 477, 2011
|
|
5MFE
 
 | | Designed armadillo repeat protein YIIIM5AII in complex with (RR)4 peptide | | Descriptor: | (RR)4, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
5MFM
 
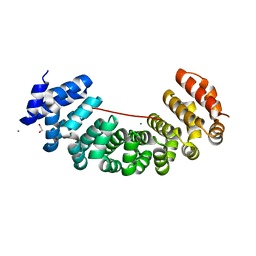 | | Designed armadillo repeat protein peptide fusion YIIIM6AII_GS11_(KR)5 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Importin subunit alpha, ... | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
7DOZ
 
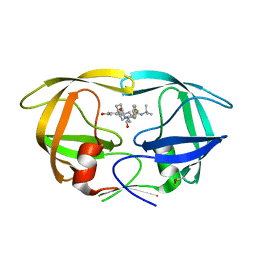 | | HIV-1 Protease D30N mutant in complex with Nelfinavir | | Descriptor: | 2-[2-HYDROXY-3-(3-HYDROXY-2-METHYL-BENZOYLAMINO)-4-PHENYL SULFANYL-BUTYL]-DECAHYDRO-ISOQUINOLINE-3-CARBOXYLIC ACID TERT-BUTYLAMIDE, Protease | | Authors: | Bihani, S.C, Hosur, M.V. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Molecular basis for reduced cleavage activity and drug resistance in D30N HIV-1 protease.
J.Biomol.Struct.Dyn., 2021
|
|
5MFL
 
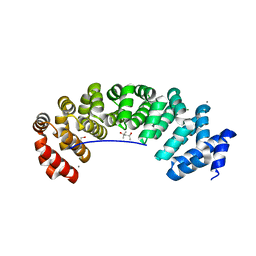 | | Designed armadillo repeat protein (KR)5_GS10_YIIIM6AII | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (KR)5_GS10_YIIIM6AII, 1,2-ETHANEDIOL, ... | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
1GO0
 
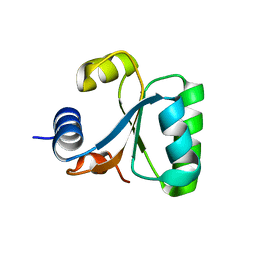 | |
5MFF
 
 | | Designed armadillo repeat protein YIIIM5AII in complex with peptide (RR)5 | | Descriptor: | (RR)5, 1,2-ETHANEDIOL, YIIIM5AII | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
5AEI
 
 | | Designed Armadillo repeat protein YIIIM5AII in complex with peptide (KR)5 | | Descriptor: | ACETATE ION, CALCIUM ION, DESIGNED ARMADILLO REPEAT PROTEIN YIIIM5AII, ... | | Authors: | Hansen, S, Tremmel, D, Madhurantakam, C, Reichen, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2015-08-31 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure and Energetic Contributions of a Designed Modular Peptide-Binding Protein with Picomolar Affinity.
J.Am.Chem.Soc., 138, 2016
|
|
5MFH
 
 | | Designed armadillo repeat protein YIIIM5AII in complex with peptide (RR)5 | | Descriptor: | (RR)5, CALCIUM ION, YIIIM5AII | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
2Z9I
 
 | |
6H45
 
 | | crystal structure of the human TGT catalytic subunit QTRT1 in complex with queuine | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Johannsson, S, Neumann, P, Ficner, R. | | Deposit date: | 2018-07-20 | | Release date: | 2018-09-05 | | Last modified: | 2023-03-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Human tRNA Guanine Transglycosylase Catalytic Subunit QTRT1.
Biomolecules, 8, 2018
|
|
5MFG
 
 | | Designed armadillo repeat protein YIIIM5AII in complex with peptide (RR)4 | | Descriptor: | (RR)4, CALCIUM ION, YIIIM5AII | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
3SPJ
 
 | |
8GWR
 
 | | Near full length Kidney type Glutaminase in complex with 2,2-Dimethyl-2,3-Dihydrobenzo[a] Phenanthridin-4(1H)-one (DDP) | | Descriptor: | 2,2-dimethyl-1,3-dihydrobenzo[a]phenanthridin-4-one, Glutaminase kidney isoform, mitochondrial | | Authors: | Shankar, S, Jobichen, C, Sivaraman, J. | | Deposit date: | 2022-09-17 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | A novel allosteric site employs a conserved inhibition mechanism in human kidney-type glutaminase.
Febs J., 290, 2023
|
|
2A0A
 
 | |
2ZFB
 
 | |
3SKX
 
 | | Crystal structure of the ATP binding domain of Archaeoglobus fulgidus COPB | | Descriptor: | ACETATE ION, Copper-exporting P-type ATPase B | | Authors: | Jayakanthan, S, Roberts, S.A, Weichsel, A, Arguello, J.M, McEvoy, M.M. | | Deposit date: | 2011-06-23 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Conformations of the apo-, substrate-bound and phosphate-bound ATP-binding domain of the Cu(II) ATPase CopB illustrate coupling of domain movement to the catalytic cycle.
Biosci.Rep., 32, 2012
|
|
5JYP
 
 | | Allosteric inhibition of Kidney Isoform of Glutaminase | | Descriptor: | 2-phenyl-~{N}-[5-[(1~{S},3~{S})-3-[5-(2-phenylethanoylamino)-1,3,4-thiadiazol-2-yl]cyclohexyl]-1,3,4-thiadiazol-2-yl]ethanamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Ramachandran, S, Sivaraman, J. | | Deposit date: | 2016-05-15 | | Release date: | 2016-08-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural basis for exploring the allosteric inhibition of human kidney type glutaminase.
Oncotarget, 7, 2016
|
|
2O8O
 
 | |
5BN5
 
 | | Structural basis for a unique ATP synthase core complex from Nanoarcheaum equitans | | Descriptor: | NEQ263, SULFATE ION, V-type ATP synthase alpha chain | | Authors: | Mohanty, S, Jobichen, C, Chichili, V.P.R, Sivaraman, J. | | Deposit date: | 2015-05-25 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.997 Å) | | Cite: | Structural Basis for a Unique ATP Synthase Core Complex from Nanoarcheaum equitans
J.Biol.Chem., 290, 2015
|
|
4PDQ
 
 | | Crystal structure of the bacterial ribosomal decoding site in complex with 4'-deoxy-4'-fluoro neomycin analog | | Descriptor: | (2S)-4-amino-N-{(1R,2S,3R,4R,5S)-5-amino-3-{[3-O-(2,6-diamino-2,6-dideoxy-beta-L-idopyranosyl)-beta-D-ribofuranosyl]oxy }-4-[(2,6-diamino-2,4,6-trideoxy-4-fluoro-alpha-D-galactopyranosyl)oxy]-2-hydroxycyclohexyl}-2-hydroxybutanamide, MAGNESIUM ION, RNA (5'-*UP*UP*GP*CP*GP*UP*CP*AP*CP*GP*CP*CP*GP*GP*CP*GP*AP*AP*GP*UP*CP*GP*C-3') | | Authors: | Hanessian, S, Saavedra, O.M, Vilchis-Reyes, M.A, Maianti, J.P, Kanazawa, H, Dozzo, P, Feeney, L.A, Armstrong, E.S, Kondo, J. | | Deposit date: | 2014-04-21 | | Release date: | 2015-01-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Synthesis, broad spectrum antibacterial activity, and X-ray co-crystal structure of the decoding bacterial ribosomal A-site with 4'-deoxy-4'-fluoro neomycin analogs
Chem Sci, 5, 2014
|
|
5NLW
 
 | | Structure of Nb36 crystal form 2 | | Descriptor: | SULFATE ION, nanobody Nb36 | | Authors: | Hansen, S.B, Andersen, K.R, Laursen, N.S, Andersen, G.R. | | Deposit date: | 2017-04-05 | | Release date: | 2017-05-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Introducing site-specific cysteines into nanobodies for mercury labelling allows de novo phasing of their crystal structures.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
