7XAD
 
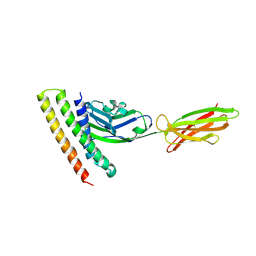 | | Crystal strucutre of PD-L1 and DBL2_02 designed protein binder | | Descriptor: | DBL2_02 binder, Programmed cell death 1 ligand 1 | | Authors: | Liu, K.F, Xu, Z.P, Han, P, Pacesa, M, Gao, G.F, Chai, Y, Tan, S.G. | | Deposit date: | 2022-03-17 | | Release date: | 2023-04-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
7XYQ
 
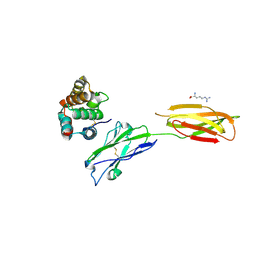 | | Crystal strucutre of PD-L1 and the computationally designed DBL1_03 protein binder | | Descriptor: | ARGININE, CD274 molecule, DBL1_03 | | Authors: | Liu, K, Xu, Z, Han, P, Pacesa, M, Gao, G.F, Chai, Y, Tan, S. | | Deposit date: | 2022-06-02 | | Release date: | 2023-04-12 | | Last modified: | 2023-05-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
3VM7
 
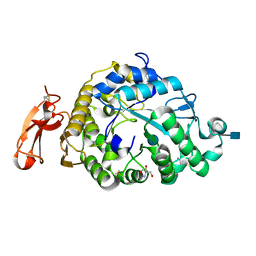 | | Structure of an Alpha-Amylase from Malbranchea cinnamomea | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-amylase, ... | | Authors: | Zhou, P, Hu, S.Q, Zhou, Y, Han, P, Yang, S.Q, Jiang, Z.Q. | | Deposit date: | 2011-12-09 | | Release date: | 2013-05-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Novel Multifunctional alpha-Amylase from the Thermophilic Fungus Malbranchea cinnamomea: Biochemical Characterization and Three-Dimensional Structure.
Appl Biochem Biotechnol., 170, 2013
|
|
7W6R
 
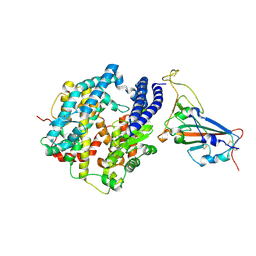 | | Structure of Bat coronavirus RaTG13 spike receptor-binding domain complexed with its receptor equine ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike glycoprotein, ... | | Authors: | Xu, Z.P, Liu, K.F, Han, P, Qi, J.X. | | Deposit date: | 2021-12-02 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Binding and structural basis of equine ACE2 to RBDs from SARS-CoV, SARS-CoV-2 and related coronaviruses
Nat Commun, 13, 2022
|
|
7W6U
 
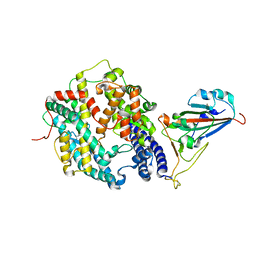 | | Structure of SARS-CoV-2 spike receptor-binding domain complexed with its receptor equine ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike protein S1, ... | | Authors: | Xu, Z.P, Liu, K.F, Han, P, Qi, J.X. | | Deposit date: | 2021-12-02 | | Release date: | 2022-06-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Binding and structural basis of equine ACE2 to RBDs from SARS-CoV, SARS-CoV-2 and related coronaviruses
Nat Commun, 13, 2022
|
|
7XBY
 
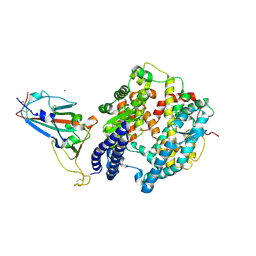 | | The crystal structure of SARS-CoV-2 Omicron BA.1 variant RBD in complex with equine ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, BROMIDE ION, ... | | Authors: | Xu, Z.P, Liu, K.F, Han, P, Qi, J.X. | | Deposit date: | 2022-03-22 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Binding and structural basis of equine ACE2 to RBDs from SARS-CoV, SARS-CoV-2 and related coronaviruses.
Nat Commun, 13, 2022
|
|
7XAE
 
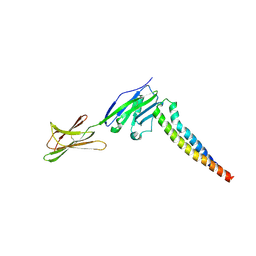 | | Crystal strucutre of PD-L1 and 3ONJA protein | | Descriptor: | 2IC6, Programmed cell death 1 ligand 1 | | Authors: | Liu, K.F, Xu, Z.P, Han, P, Gao, G.F, Chai, Y, Tan, S.G. | | Deposit date: | 2022-03-17 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.44 Å) | | Cite: | Crystal strucutre of PD-L1 and 2IC6 protein
To Be Published
|
|
7XBG
 
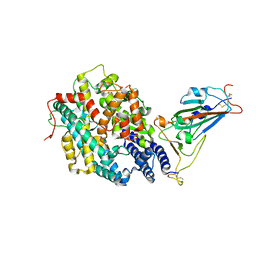 | | The crystal structure of RshSTT182/200 RBD-insert2-T346R-Y496G mutant in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Hu, Y, Liu, K.F, Han, P, Qi, J.X. | | Deposit date: | 2022-03-21 | | Release date: | 2023-01-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.37 Å) | | Cite: | Host range and structural analysis of bat-origin RshSTT182/200 coronavirus binding to human ACE2 and its animal orthologs.
Embo J., 42, 2023
|
|
7XBH
 
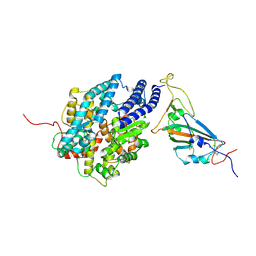 | | The complex structure of RshSTT182/200 RBD bound to human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, RshSTT182/200 coronavirus receptor binding domain, ... | | Authors: | Hu, Y, Liu, K.F, Han, P, Qi, J.X. | | Deposit date: | 2022-03-21 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Host range and structural analysis of bat-origin RshSTT182/200 coronavirus binding to human ACE2 and its animal orthologs.
Embo J., 42, 2023
|
|
7XBF
 
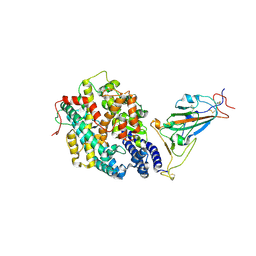 | | The complex structure of RshSTT182/200 RBD-insert2 bound to human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, RshSTT182/200 coronavirus receptor binding domain insert2, ... | | Authors: | Hu, Y, Liu, K.F, Han, P, Qi, J.X. | | Deposit date: | 2022-03-21 | | Release date: | 2023-01-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Host range and structural analysis of bat-origin RshSTT182/200 coronavirus binding to human ACE2 and its animal orthologs.
Embo J., 42, 2023
|
|
4XHR
 
 | | Structure of a phospholipid trafficking complex, native | | Descriptor: | Mitochondrial distribution and morphology protein 35, Protein UPS1, mitochondrial | | Authors: | Yu, F, He, F, Wang, C, Zhang, P. | | Deposit date: | 2015-01-06 | | Release date: | 2015-07-01 | | Last modified: | 2015-08-05 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis of intramitochondrial phosphatidic acid transport mediated by Ups1-Mdm35 complex
Embo Rep., 16, 2015
|
|
4XIZ
 
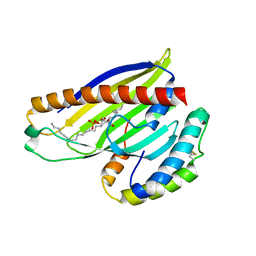 | | Structure of a phospholipid trafficking complex with substrate | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, Mitochondrial distribution and morphology protein 35, Protein UPS1, ... | | Authors: | Yu, F, He, F, Wang, C, Zhang, P. | | Deposit date: | 2015-01-08 | | Release date: | 2015-07-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of intramitochondrial phosphatidic acid transport mediated by Ups1-Mdm35 complex
Embo Rep., 16, 2015
|
|
7WSE
 
 | |
7WSF
 
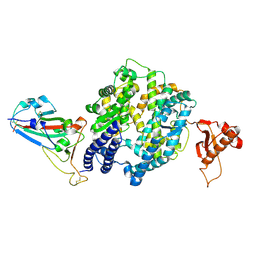 | |
7WSH
 
 | | Cryo-EM structure of SARS-CoV-2 spike receptor-binding domain in complex with sea lion ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike protein S1, ... | | Authors: | Li, S, Han, P, Qi, J. | | Deposit date: | 2022-01-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Cross-species recognition and molecular basis of SARS-CoV-2 and SARS-CoV binding to ACE2s of marine animals.
Natl Sci Rev, 9, 2022
|
|
7WSG
 
 | |
4XOK
 
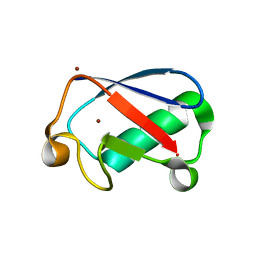 | | Observing the overall rocking motion of a protein in a crystal. | | Descriptor: | Ubiquitin, ZINC ION | | Authors: | Coquelle, N, Ma, P, Schanda, P, Colletier, J.P. | | Deposit date: | 2015-01-16 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Observing the overall rocking motion of a protein in a crystal.
Nat Commun, 6, 2015
|
|
6LZ7
 
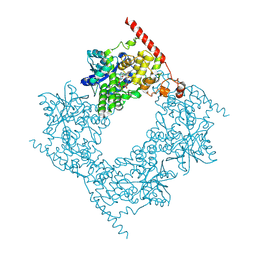 | |
6LZ3
 
 | |
6SKM
 
 | | Structure of the native full-length HIV-1 capsid protein A92E in helical assembly (-13,12) | | Descriptor: | Gag protein | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2019-08-16 | | Release date: | 2020-08-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6SLQ
 
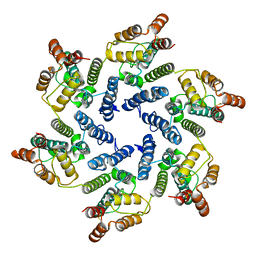 | | Structure of the native full-length HIV-1 capsid protein A92E in helical assembly (-12,11) | | Descriptor: | Gag protein | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2019-08-20 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6SLU
 
 | | Structure of the native full-length HIV-1 capsid protein A92E in helical assembly (-13,11) | | Descriptor: | Gag protein | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2019-08-20 | | Release date: | 2020-09-09 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6SMU
 
 | | Structure of the native full-length HIV-1 capsid protein in helical assembly (-13,12) | | Descriptor: | Gag protein | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2019-08-22 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6Q0X
 
 | |
6XF8
 
 | | DLP 5 fold | | Descriptor: | Inner capsid protein lambda-1, Inner capsid protein sigma-2, Outer capsid protein mu-1, ... | | Authors: | Sutton, G, Sun, D.P, Fu, X.F, Kotecha, A, Hecksel, G.W, Clare, D.K, Zhang, P, Stuart, D, Boyce, M. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Assembly intermediates of orthoreovirus captured in the cell.
Nat Commun, 11, 2020
|
|
