6KMM
 
 | | Crystal Structure of HEPES bound Dye Decolorizing peroxidase from Bacillus subtilis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Dhankhar, P, Dalal, V, Mahto, J.K, Kumar, P. | | Deposit date: | 2019-07-31 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Characterization of dye-decolorizing peroxidase from Bacillus subtilis.
Arch.Biochem.Biophys., 693, 2020
|
|
6KMN
 
 | | Crystal Structure of Dye Decolorizing peroxidase from Bacillus subtilis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Deferrochelatase/peroxidase EfeB, ... | | Authors: | Dhankhar, P, Dalal, V, Mahto, J.K, Kumar, P. | | Deposit date: | 2019-07-31 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Characterization of dye-decolorizing peroxidase from Bacillus subtilis.
Arch.Biochem.Biophys., 693, 2020
|
|
5LC3
 
 | | Xray structure of mouse FAM3C ILEI monomer | | Descriptor: | Protein FAM3C | | Authors: | Johansson, P, Jansson, A. | | Deposit date: | 2016-06-19 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The interleukin-like epithelial-mesenchymal transition inducer ILEI exhibits a non-interleukin-like fold and is active as a domain-swapped dimer.
J. Biol. Chem., 292, 2017
|
|
5LC2
 
 | | Xray structure of human FAM3C ILEI monomer | | Descriptor: | Protein FAM3C | | Authors: | Johansson, P, Jansson, A. | | Deposit date: | 2016-06-19 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The interleukin-like epithelial-mesenchymal transition inducer ILEI exhibits a non-interleukin-like fold and is active as a domain-swapped dimer.
J. Biol. Chem., 292, 2017
|
|
5LC4
 
 | | Xray structure of mouse FAM3C ILEI dimer | | Descriptor: | Protein FAM3C | | Authors: | Johansson, P, Jansson, A. | | Deposit date: | 2016-06-19 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The interleukin-like epithelial-mesenchymal transition inducer ILEI exhibits a non-interleukin-like fold and is active as a domain-swapped dimer.
J. Biol. Chem., 292, 2017
|
|
1DRO
 
 | | NMR STRUCTURE OF THE CYTOSKELETON/SIGNAL TRANSDUCTION PROTEIN | | Descriptor: | BETA-SPECTRIN | | Authors: | Zhang, P, Talluri, S, Deng, H, Branton, D, Wagner, G. | | Deposit date: | 1995-09-29 | | Release date: | 1996-04-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the pleckstrin homology domain of Drosophila beta-spectrin.
Structure, 3, 1995
|
|
7FHA
 
 | | Crystal structure of the ATP sulfurylase domain of human PAPSS2 in complex with APS | | Descriptor: | ADENOSINE-5'-PHOSPHOSULFATE, Bifunctional 3'-phosphoadenosine 5'-phosphosulfate synthase 2, POTASSIUM ION, ... | | Authors: | Zhang, P, Zhang, L, Zhang, L. | | Deposit date: | 2021-07-29 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the substrate recognition mechanism of ATP-sulfurylase domain of human PAPS synthase 2.
Biochem.Biophys.Res.Commun., 586, 2022
|
|
7FH3
 
 | | Crystal structure of the ATP sulfurylase domain of human PAPSS2 | | Descriptor: | Bifunctional 3'-phosphoadenosine 5'-phosphosulfate synthase 2, SULFATE ION, beta-D-glucopyranose | | Authors: | Zhang, P, Zhang, L, Zhang, L. | | Deposit date: | 2021-07-29 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the substrate recognition mechanism of ATP-sulfurylase domain of human PAPS synthase 2.
Biochem.Biophys.Res.Commun., 586, 2022
|
|
1M0V
 
 | | NMR STRUCTURE OF THE TYPE III SECRETORY DOMAIN OF YERSINIA YOPH COMPLEXED WITH THE SKAP-HOM PHOSPHO-PEPTIDE N-acetyl-DEpYDDPF-NH2 | | Descriptor: | PROTEIN-TYROSINE PHOSPHATASE YOPH, SKAP55 homologue | | Authors: | Khandelwal, P, Keliikuli, K, Smith, C.L, Saper, M.A, Zuiderweg, E.R.P. | | Deposit date: | 2002-06-14 | | Release date: | 2002-07-24 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure and phosphopeptide binding to the N-terminal domain of Yersinia YopH: comparison with a crystal structure
Biochemistry, 41, 2002
|
|
7DLK
 
 | | Crystal Structure of veratryl alcohol bound Dye Decolorizing peroxidase from Bacillus subtilis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Dhankhar, P, Dalal, V, Kumar, P. | | Deposit date: | 2020-11-27 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of dye-decolorizing peroxidase from Bacillus subtilis in complex with veratryl alcohol.
Int.J.Biol.Macromol., 193, 2021
|
|
7E5Q
 
 | | Crystal Structure of Dye Decolorizing peroxidase from Bacillus subtilis at acidic pH | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Dhankhar, P, Dalal, V, Kumar, P. | | Deposit date: | 2021-02-19 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights at acidic pH of dye-decolorizing peroxidase from Bacillus subtilis.
Proteins, 2022
|
|
5EB8
 
 | |
5EBA
 
 | |
3P5N
 
 | |
5EFD
 
 | | Crystal structure of a surface pocket creating mutant (W6A) of an alkali thermostable GH10 xylanase from Bacillus sp. NG-27 | | Descriptor: | 1,2-ETHANEDIOL, Beta-xylanase, CHLORIDE ION, ... | | Authors: | Mahanta, P, Bhardwaj, A, Reddy, V.S, Ramakumar, S. | | Deposit date: | 2015-10-23 | | Release date: | 2016-10-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.674 Å) | | Cite: | Small Glycols Discover Cryptic Pockets on Proteins for Fragment-Based Approaches.
J.Chem.Inf.Model., 2021
|
|
8P5O
 
 | | Proline activating adenylation domain of gramicidin S synthetase 2 - GrsB1-Acore | | Descriptor: | Gramicidin S synthase 2 | | Authors: | Stephan, P, Basquin, J, Caputi, L, O'Connor, S.E, Kries, H. | | Deposit date: | 2023-05-24 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Directed Evolution of Piperazic Acid Incorporation by a Nonribosomal Peptide Synthetase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
5EFF
 
 | | Crystal structure of an aromatic mutant (F4A) of an alkali thermostable GH10 xylanase from Bacillus sp. NG-27 | | Descriptor: | Beta-xylanase, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Mahanta, P, Bhardwaj, A, Reddy, V.S, Ramakumar, S. | | Deposit date: | 2015-10-23 | | Release date: | 2016-10-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal structure of an aromatic mutant (F4A) of an alkali thermostable GH10 xylanase from Bacillus sp. NG-27
To Be Published
|
|
4HUQ
 
 | | Crystal Structure of a transporter | | Descriptor: | Energy-coupling factor transporter ATP-binding protein EcfA 1, Energy-coupling factor transporter ATP-binding protein EcfA 2, Energy-coupling factor transporter transmembrane protein EcfT, ... | | Authors: | Zhang, P, Xu, K, Zhang, M, Zhao, Q, Yu, F. | | Deposit date: | 2012-11-03 | | Release date: | 2013-04-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.998 Å) | | Cite: | Crystal structure of a folate energy-coupling factor transporter from Lactobacillus brevis.
Nature, 497, 2013
|
|
4GHU
 
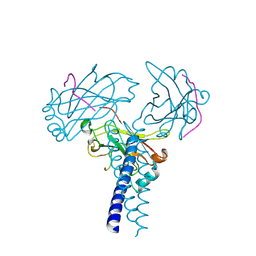 | | Crystal structure of TRAF3/Cardif | | Descriptor: | Mitochondrial antiviral-signaling protein, TNF receptor-associated factor 3 | | Authors: | Zhang, P. | | Deposit date: | 2012-08-08 | | Release date: | 2012-11-28 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Single Amino Acid Substitutions Confer the Antiviral Activity of the TRAF3 Adaptor Protein onto TRAF5
Sci.Signal., 5, 2012
|
|
3HMJ
 
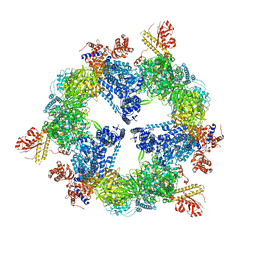 | | Saccharomyces cerevisiae FAS type I | | Descriptor: | (2S, 3R)-3-HYDROXY-4-OXO-7,10-TRANS,TRANS-DODECADIENAMIDE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Johansson, P, Mulinacci, B, Koestler, C, Vollrath, R, Oesterhelt, D, Grininger, M. | | Deposit date: | 2009-05-29 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Multimeric options for the auto-activation of the Saccharomyces cerevisiae FAS type I megasynthase
Structure, 17, 2009
|
|
5JNQ
 
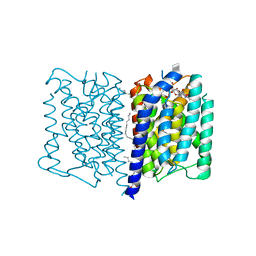 | | MraY tunicamycin complex | | Descriptor: | PALMITIC ACID, Phospho-N-acetylmuramoyl-pentapeptide-transferase, Tunicamycin, ... | | Authors: | Johansson, P. | | Deposit date: | 2016-04-30 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | MraY-antibiotic complex reveals details of tunicamycin mode of action.
Nat. Chem. Biol., 13, 2017
|
|
2VKZ
 
 | | Structure of the cerulenin-inhibited fungal fatty acid synthase type I multienzyme complex | | Descriptor: | (2S, 3R)-3-HYDROXY-4-OXO-7,10-TRANS,TRANS-DODECADIENAMIDE, FATTY ACID SYNTHASE SUBUNIT ALPHA, ... | | Authors: | Johansson, P, Wiltschi, B, Kumari, P, Kessler, B, Vonrhein, C, Vonck, J, Oesterhelt, D, Grininger, M. | | Deposit date: | 2008-01-07 | | Release date: | 2008-08-12 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Inhibition of the Fungal Fatty Acid Synthase Type I Multienzyme Complex.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
4GJH
 
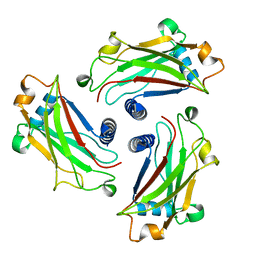 | | Crystal Structure of the TRAF domain of TRAF5 | | Descriptor: | TNF receptor-associated factor 5 | | Authors: | Zhang, P, Reichardt, A, Liang, H, Wang, Y, Cheng, D, Aliyari, R, Cheng, G, Liu, Y. | | Deposit date: | 2012-08-09 | | Release date: | 2012-11-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Single Amino Acid Substitutions Confer the Antiviral Activity of the TRAF3 Adaptor Protein onto TRAF5
Sci.Signal., 5, 2012
|
|
3J82
 
 | | Electron cryo-microscopy of DNGR-1 in complex with F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Hanc, P, Fujii, T, Yamada, Y, Huotari, J, Schulz, O, Ahrens, S, Kjaer, S, Way, M, Namba, K, Reis e Sousa, C. | | Deposit date: | 2014-09-25 | | Release date: | 2015-05-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structure of the Complex of F-Actin and DNGR-1, a C-Type Lectin Receptor Involved in Dendritic Cell Cross-Presentation of Dead Cell-Associated Antigens.
Immunity, 42, 2015
|
|
2F5K
 
 | |
