7ZVR
 
 | | Crystal structure of the carotenoid-binding protein domain from silkworm Bombyx mori (BmCBP) complexed with zeaxanthin | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, Carotenoid-binding protein | | Authors: | Sluchanko, N.N, Boyko, K.M, Varfolomeeva, L.A, Slonimskiy, Y.B, Egorkin, N.A, Maksimov, E.G, Popov, V.O. | | Deposit date: | 2022-05-17 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the carotenoid binding and transport function of a START domain.
Structure, 30, 2022
|
|
7ZTU
 
 | | Crystal structure of the carotenoid-binding protein domain from silkworm Bombyx mori (BmCBP) in the apoform, D162L mutant | | Descriptor: | Carotenoid-binding protein | | Authors: | Sluchanko, N.N, Boyko, K.M, Varfolomeeva, L.A, Slonimskiy, Y.B, Egorkin, N.A, Maksimov, E.G, Popov, V.O. | | Deposit date: | 2022-05-11 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Silkworm carotenoprotein as an efficient carotenoid extractor, solubilizer and transporter.
Int.J.Biol.Macromol., 223, 2022
|
|
7ZVQ
 
 | | Crystal structure of the carotenoid-binding protein domain from silkworm Bombyx mori (BmCBP) in the apoform, S206V mutant | | Descriptor: | Carotenoid-binding protein | | Authors: | Sluchanko, N.N, Boyko, K.M, Varfolomeeva, L.A, Slonimskiy, Y.B, Egorkin, N.A, Maksimov, E.G, Popov, V.O. | | Deposit date: | 2022-05-17 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the carotenoid binding and transport function of a START domain.
Structure, 30, 2022
|
|
7ZTQ
 
 | | Crystal structure of the carotenoid-binding protein domain from silkworm Bombyx mori (BmCBP) in the apoform | | Descriptor: | Carotenoid-binding protein, GLYCEROL | | Authors: | Sluchanko, N.N, Boyko, K.M, Varfolomeeva, L.A, Slonimskiy, Y.B, Egorkin, N.A, Maksimov, E.G, Popov, V.O. | | Deposit date: | 2022-05-11 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Silkworm carotenoprotein as an efficient carotenoid extractor, solubilizer and transporter.
Int.J.Biol.Macromol., 223, 2022
|
|
1S8K
 
 | | Solution Structure of BmKK4, A Novel Potassium Channel Blocker from Scorpion Buthus martensii Karsch, 25 structures | | Descriptor: | Toxin BmKK4 | | Authors: | Zhang, N, Chen, X, Li, M, Cao, C, Wang, Y, Hu, G, Wu, H. | | Deposit date: | 2004-02-02 | | Release date: | 2005-02-08 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BmKK4, the first member of subfamily alpha-KTx 17 of scorpion toxins
Biochemistry, 43, 2004
|
|
2OEY
 
 | | Solution Structure of a Designed Spirocyclic Helical Ligand Binding at a Two-Base Bulge Site in DNA | | Descriptor: | (1R,3A'S,10'S,10A'R)-7-METHOXY-2-OXO-10',10A'-DIHYDRO-2H,3A'H-SPIRO[NAPHTHALENE-1,3'-PENTALENO[1,2-B]NAPHTHALEN]-10'-YL 2,6-DIDEOXY-2-(METHYLAMINO)-ALPHA-D-GALACTOPYRANOSIDE, DNA (25-MER) | | Authors: | Zhang, N, Lin, Y, Xiao, Z, Jones, G.B, Goldberg, I.H. | | Deposit date: | 2007-01-01 | | Release date: | 2007-04-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Designed Spirocyclic Helical Ligand Binding at a Two-Base Bulge Site in DNA.
Biochemistry, 46, 2007
|
|
7WGI
 
 | | Crystal structure of AflSQS from Aspergillus flavus | | Descriptor: | INDOLE, PHOSPHATE ION, Squalene synthase | | Authors: | Shang, N, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2021-12-28 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Structural and Bioinformatics Investigation of a Fungal Squalene Synthase and Comparisons with Other Membrane Proteins.
Acs Omega, 7, 2022
|
|
7WGH
 
 | | Crystal structure of AflSQS from Aspergillus flavus in complex with FSPP | | Descriptor: | PHOSPHATE ION, PYROPHOSPHATE 2-, S-[(2E,6E)-3,7,11-TRIMETHYLDODECA-2,6,10-TRIENYL] TRIHYDROGEN THIODIPHOSPHATE, ... | | Authors: | Shang, N, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2021-12-28 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | A Structural and Bioinformatics Investigation of a Fungal Squalene Synthase and Comparisons with Other Membrane Proteins.
Acs Omega, 7, 2022
|
|
7L5M
 
 | | Crystal Structure of the DiB-RM-split Protein | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lipocalin family protein, SODIUM ION, ... | | Authors: | Bozhanova, N.G, Harp, J.M, Meiler, J. | | Deposit date: | 2020-12-22 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Computational redesign of a fluorogen activating protein with Rosetta.
Plos Comput.Biol., 17, 2021
|
|
7L5L
 
 | |
7L5K
 
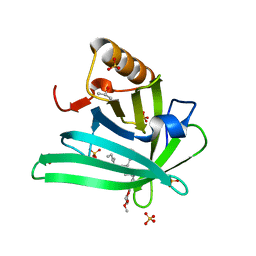 | | Crystal structure of the DiB-RM protein | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, ISOPROPYL ALCOHOL, Lipocalin family protein, ... | | Authors: | Bozhanova, N.G, Harp, J.M, Meiler, J. | | Deposit date: | 2020-12-22 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Computational redesign of a fluorogen activating protein with Rosetta.
Plos Comput.Biol., 17, 2021
|
|
7LG4
 
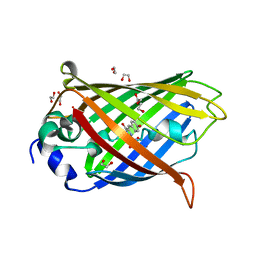 | |
1Y9H
 
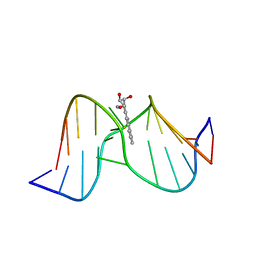 | | Methylation of cytosine at C5 in a CpG sequence context causes a conformational switch of a benzo[a]pyrene diol epoxide-N2-guanine adduct in DNA from a minor groove alignment to intercalation with base displacement | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, 5'-D(*CP*CP*AP*TP*(5CM)P*(BPG)P*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3' | | Authors: | Zhang, N, Lin, C, Huang, X, Kolbanovskiy, A, Hingerty, B.E, Amin, S, Broyde, S, Geacintov, N.E, Patel, D.J. | | Deposit date: | 2004-12-15 | | Release date: | 2005-03-22 | | Last modified: | 2024-04-24 | | Method: | SOLUTION NMR | | Cite: | Methylation of cytosine at C5 in a CpG sequence context causes a conformational switch of a benzo[a]pyrene diol epoxide-N2-guanine adduct in DNA from a minor groove alignment to intercalation with base displacement.
J.Mol.Biol., 346, 2005
|
|
7M69
 
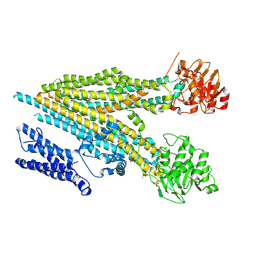 | |
7M68
 
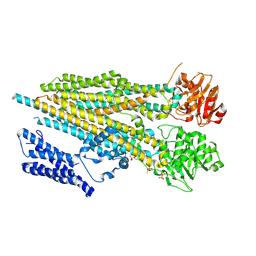 | |
7O5H
 
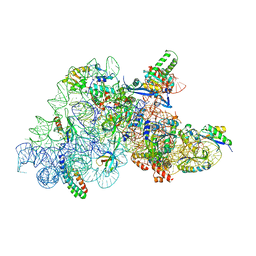 | | Ribosomal methyltransferase KsgA bound to small ribosomal subunit | | Descriptor: | 16S rRNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Stephan, N.C, Ries, A.B, Boehringer, D, Ban, N. | | Deposit date: | 2021-04-08 | | Release date: | 2021-06-16 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of successive adenosine modifications by the conserved ribosomal methyltransferase KsgA.
Nucleic Acids Res., 49, 2021
|
|
7KS0
 
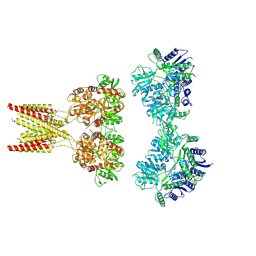 | | GluK2/K5 with 6-Cyano-7-nitroquinoxaline-2,3-dione (CNQX) | | Descriptor: | Glutamate receptor ionotropic, kainate 2, kainate 5,Green fluorescent protein chimera | | Authors: | Khanra, N, Brown, P.M.G.E, Perozzo, A.M, Bowie, D, Meyerson, J.R. | | Deposit date: | 2020-11-20 | | Release date: | 2021-03-24 | | Last modified: | 2021-07-07 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Architecture and structural dynamics of the heteromeric GluK2/K5 kainate receptor.
Elife, 10, 2021
|
|
7KS3
 
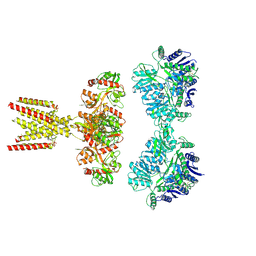 | | GluK2/K5 with L-Glu | | Descriptor: | Glutamate receptor ionotropic, kainate 2, kainate 5,Green fluorescent protein chimera | | Authors: | Khanra, N, Brown, P.M.G.E, Perozzo, A.M, Bowie, D, Meyerson, J.R. | | Deposit date: | 2020-11-20 | | Release date: | 2021-03-24 | | Last modified: | 2021-07-07 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Architecture and structural dynamics of the heteromeric GluK2/K5 kainate receptor.
Elife, 10, 2021
|
|
1UAN
 
 | | Crystal structure of the conserved protein TT1542 from Thermus thermophilus HB8 | | Descriptor: | hypothetical protein TT1542 | | Authors: | Handa, N, Terada, T, Tame, J.R.H, Park, S.-Y, Kinoshita, K, Ota, M, Nakamura, H, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-03-12 | | Release date: | 2003-08-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the conserved protein TT1542 from Thermus thermophilus HB8
PROTEIN SCI., 12, 2003
|
|
1B7F
 
 | | SXL-LETHAL PROTEIN/RNA COMPLEX | | Descriptor: | PROTEIN (SXL-LETHAL PROTEIN), RNA (5'-R(P*GP*UP*UP*GP*UP*UP*UP*UP*UP*UP*UP*U)-3') | | Authors: | Handa, N, Nureki, O, Kurimoto, K, Kim, I, Sakamoto, H, Shimura, Y, Muto, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-01-23 | | Release date: | 1999-05-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein.
Nature, 398, 1999
|
|
3U82
 
 | | Binding of herpes simplex virus glycoprotein D to nectin-1 exploits host cell adhesion | | Descriptor: | Envelope glycoprotein D, Poliovirus receptor-related protein 1 | | Authors: | Zhang, N, Yan, J, Lu, G, Guo, Z, Fan, Z, Wang, J, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2011-10-15 | | Release date: | 2012-03-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.164 Å) | | Cite: | Binding of herpes simplex virus glycoprotein D to nectin-1 exploits host cell adhesion.
Nat Commun, 2, 2011
|
|
1BUW
 
 | |
1HIV
 
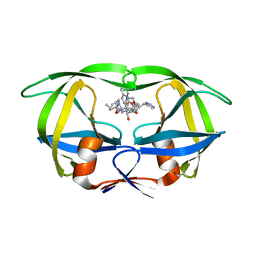 | | CRYSTAL STRUCTURE OF A COMPLEX OF HIV-1 PROTEASE WITH A DIHYDROETHYLENE-CONTAINING INHIBITOR: COMPARISONS WITH MOLECULAR MODELING | | Descriptor: | 4-[(2R)-3-{[(1S,2S,3R,4S)-1-(cyclohexylmethyl)-2,3-dihydroxy-5-methyl-4-({(1S,2R)-2-methyl-1-[(pyridin-2-ylmethyl)carbamoyl]butyl}carbamoyl)hexyl]amino}-2-{[(naphthalen-1-yloxy)acetyl]amino}-3-oxopropyl]-1H-imidazol-3-ium, HIV-1 PROTEASE | | Authors: | Thanki, N, Wlodawer, A. | | Deposit date: | 1992-02-12 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a complex of HIV-1 protease with a dihydroxyethylene-containing inhibitor: comparisons with molecular modeling.
Protein Sci., 1, 1992
|
|
2Q89
 
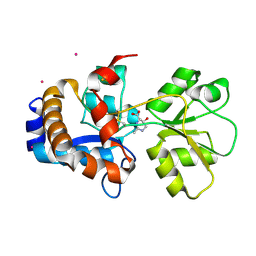 | | Crystal structure of EhuB in complex with hydroxyectoine | | Descriptor: | (4S,5S)-5-HYDROXY-2-METHYL-1,4,5,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, CADMIUM ION, Putative ABC transporter amino acid-binding protein | | Authors: | Hanekop, N, Hoeing, M, Sohn-Bosser, L, Jebbar, M, Schmitt, L, Bremer, E. | | Deposit date: | 2007-06-09 | | Release date: | 2008-01-01 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the ligand-binding protein EhuB from Sinorhizobium meliloti reveals substrate recognition of the compatible solutes ectoine and hydroxyectoine.
J.Mol.Biol., 374, 2007
|
|
1WMG
 
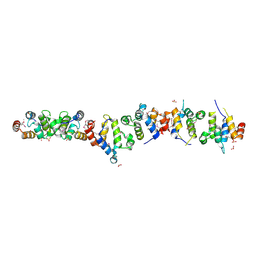 | | Crystal structure of the UNC5H2 death domain | | Descriptor: | SULFATE ION, SULFITE ION, netrin receptor Unc5h2 | | Authors: | Handa, N, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-07-09 | | Release date: | 2005-01-09 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the UNC5H2 death domain
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
