4ZUU
 
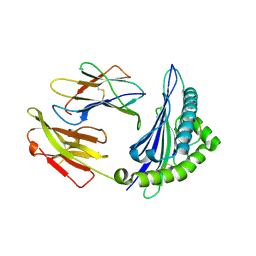 | | Crystal structure of Equine MHC I(Eqca-N*00602) in complexed with equine infectious anaemia virus (EIAV) derived peptide Gag-CF9 | | Descriptor: | Beta-2-microglobulin, CYS-THR-SER-GLU-GLU-MET-ASN-ALA-PHE, Classical MHC class I antigen | | Authors: | Yao, S, Liu, J, Qi, J, Chen, R, Zhang, N, Liu, Y, Xia, C. | | Deposit date: | 2015-05-17 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Illumination of Equine MHC Class I Molecules Highlights Unconventional Epitope Presentation Manner That Is Evolved in Equine Leukocyte Antigen Alleles
J Immunol., 196, 2016
|
|
6S67
 
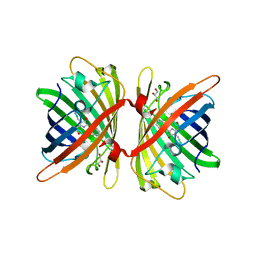 | | Structure of the Fluorescent Protein AausFP1 from Aequorea cf. australis at pH 7.0 | | Descriptor: | Aequorea cf. australis fluorescent protein 1 (AausFP1), GLYCEROL | | Authors: | Depernet, H, Gotthard, G, Lambert, G.G, Shaner, N.C, Royant, A. | | Deposit date: | 2019-07-02 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Aequorea's secrets revealed: New fluorescent proteins with unique properties for bioimaging and biosensing.
Plos Biol., 18, 2020
|
|
5Y86
 
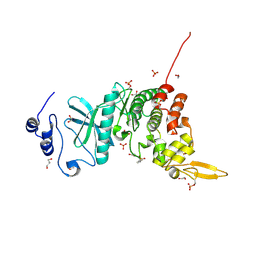 | | Crystal structure of kinase | | Descriptor: | 1,2-ETHANEDIOL, 7-METHOXY-1-METHYL-9H-BETA-CARBOLINE, Dual specificity tyrosine-phosphorylation-regulated kinase 3, ... | | Authors: | Kim, K.L, Cha, J.S, Cho, Y.S, Kim, H.Y, Chang, N.P, Cho, H.S. | | Deposit date: | 2017-08-18 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Human Dual-Specificity Tyrosine-Regulated Kinase 3 Reveals New Structural Features and Insights into its Auto-phosphorylation
J. Mol. Biol., 430, 2018
|
|
4ZUT
 
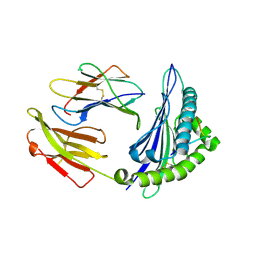 | | Crystal structure of Equine MHC I(Eqca-N*00602) in complexed with equine infectious anaemia virus (EIAV) derived peptide Gag-GW12 | | Descriptor: | Beta-2-microglobulin, Classical MHC class I antigen, GLY-SER-GLN-LYS-LEU-THR-THR-GLY-ASN-CYS-ASN-TRP | | Authors: | Yao, S, Liu, J, Qi, J, Chen, R, Zhang, N, Liu, Y, Xia, C. | | Deposit date: | 2015-05-17 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Illumination of Equine MHC Class I Molecules Highlights Unconventional Epitope Presentation Manner That Is Evolved in Equine Leukocyte Antigen Alleles
J Immunol., 196, 2016
|
|
3AQS
 
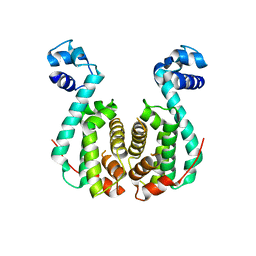 | | Crystal structure of RolR (NCGL1110) without ligand | | Descriptor: | Bacterial regulatory proteins, tetR family | | Authors: | Li, D.F, Zhang, N, Hou, Y.J, Liu, S.J, Wang, D.C. | | Deposit date: | 2010-11-18 | | Release date: | 2011-07-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal structures of the transcriptional repressor RolR reveals a novel recognition mechanism between inducer and regulator.
Plos One, 6, 2011
|
|
4ZUS
 
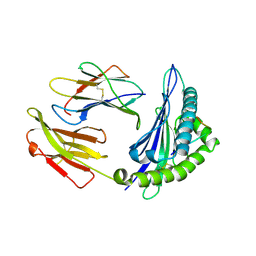 | | Crystal structure of Equine MHC I(Eqca-N*00602) in complexed with equine infectious anaemia virus (EIAV) derived peptide REV-QW11 | | Descriptor: | Beta-2-microglobulin, Classical MHC class I antigen, GLN-ALA-GLU-VAL-LEU-GLN-GLU-ARG-LEU-GLU-TRP | | Authors: | Yao, S, Liu, J, Qi, J, Chen, R, Zhang, N, Liu, Y, Xia, C. | | Deposit date: | 2015-05-17 | | Release date: | 2016-04-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Illumination of Equine MHC Class I Molecules Highlights Unconventional Epitope Presentation Manner That Is Evolved in Equine Leukocyte Antigen Alleles
J Immunol., 196, 2016
|
|
4ZUW
 
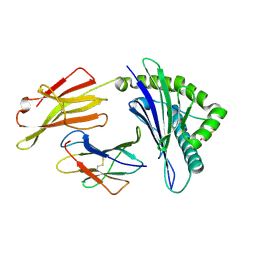 | | Crystal structure of Equine MHC I(Eqca-N*00601) in complexed with equine infectious anaemia virus (EIAV) derived peptide Gag-GW12 | | Descriptor: | Beta-2-microglobulin, Classical MHC class I antigen, GLY-SER-GLN-LYS-LEU-THR-THR-GLY-ASN-CYS-ASN-TRP | | Authors: | Yao, S, Liu, J, Qi, J, Chen, R, Zhang, N, Liu, Y, Xia, C. | | Deposit date: | 2015-05-17 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Illumination of Equine MHC Class I Molecules Highlights Unconventional Epitope Presentation Manner That Is Evolved in Equine Leukocyte Antigen Alleles
J Immunol., 196, 2016
|
|
6S68
 
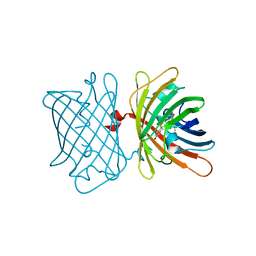 | | Structure of the Fluorescent Protein AausFP2 from Aequorea cf. australis at pH 7.6 | | Descriptor: | Aequorea cf. australis fluorescent protein 2 (AausFP2) | | Authors: | Depernet, H, Gotthard, G, Lambert, G.G, Shaner, N.C, Royant, A. | | Deposit date: | 2019-07-02 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Aequorea's secrets revealed: New fluorescent proteins with unique properties for bioimaging and biosensing.
Plos Biol., 18, 2020
|
|
3AQT
 
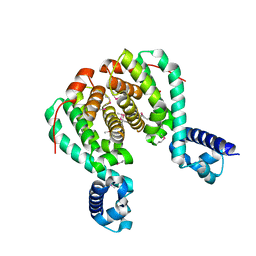 | | CRYSTAL STRUCTURE OF RolR (NCGL1110) complex WITH ligand RESORCINOL | | Descriptor: | Bacterial regulatory proteins, tetR family, RESORCINOL | | Authors: | Li, D.F, Zhang, N, Hou, Y.J, Liu, S.J, Wang, D.C. | | Deposit date: | 2010-11-18 | | Release date: | 2011-07-06 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the transcriptional repressor RolR reveals a novel recognition mechanism between inducer and regulator.
Plos One, 6, 2011
|
|
1EET
 
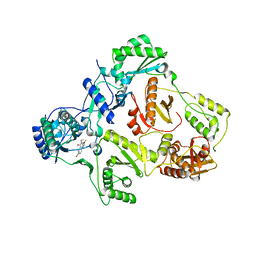 | | HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH THE INHIBITOR MSC204 | | Descriptor: | 1-(5-BROMO-PYRIDIN-2-YL)-3-[2-(6-FLUORO-2-HYDROXY-3-PROPIONYL-PHENYL)-CYCLOPROPYL]-UREA, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Hogberg, M, Sahlberg, C, Engelhardt, P, Noreen, R, Kangasmetsa, J, Johansson, N.G, Oberg, B, Vrang, L, Zhang, H, Sahlberg, B.L, Unge, T, Lovgren, S, Fridborg, K, Backbro, K. | | Deposit date: | 2000-02-03 | | Release date: | 2001-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Urea-PETT compounds as a new class of HIV-1 reverse transcriptase inhibitors. 3. Synthesis and further structure-activity relationship studies of PETT analogues.
J.Med.Chem., 42, 1999
|
|
5HL0
 
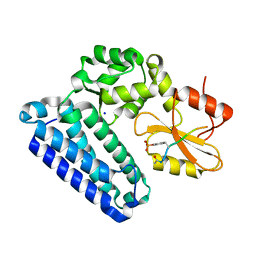 | | Crystal Structure of c-Cbl TKBD in complex with SPRY2 peptide (54-60, pY55) Refined to 2.2A Resolution | | Descriptor: | E3 ubiquitin-protein ligase CBL, SODIUM ION, Sprouty 2 (SPRY2) | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of c-Cbl TKBD in complex with SPRY2 peptide (54-60, pY55) Refined to 2.2A Resolution
To Be Published
|
|
5HKX
 
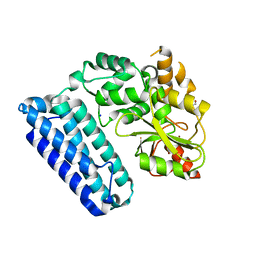 | | Crystal Structure of c-Cbl TKBD-RING domains (Y371E mutant) Refined to 1.85 A Resolution | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase CBL, SODIUM ION, ... | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of c-Cbl TKBD-RING domains (Y371E mutant) Refined to 1.85 A Resolution
To be published
|
|
7M5Z
 
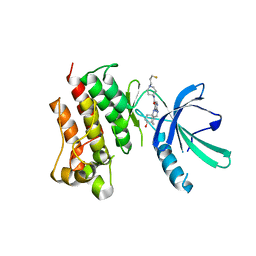 | | Crystal Structure of the MerTK Kinase Domain in Complex with Inhibitor MIPS15692 | | Descriptor: | 2-(butylamino)-N-[1-(3-fluoropropyl)piperidin-4-yl]-4-{[(1r,4r)-4-hydroxycyclohexyl]amino}pyrimidine-5-carboxamide, Tyrosine-protein kinase Mer | | Authors: | Hermans, S.J, Hancock, N.C, Baell, J.B, Parker, M.W. | | Deposit date: | 2021-03-25 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Development of [ 18 F]MIPS15692, a radiotracer with in vitro proof-of-concept for the imaging of MER tyrosine kinase (MERTK) in neuroinflammatory disease.
Eur.J.Med.Chem., 226, 2021
|
|
3WJA
 
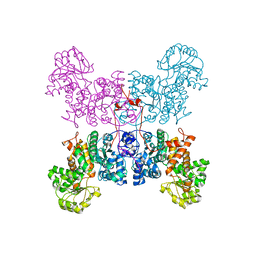 | | The crystal structure of human cytosolic NADP(+)-dependent malic enzyme in apo form | | Descriptor: | NADP-dependent malic enzyme | | Authors: | Li, S.-Y, Chen, M.-C, Yang, P.-C, Chan, N.-L, Liu, J.-H, Hung, H.-C. | | Deposit date: | 2013-10-08 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.548 Å) | | Cite: | Structural characteristics of the nonallosteric human cytosolic malic enzyme.
Biochim.Biophys.Acta, 1844, 2014
|
|
5HKZ
 
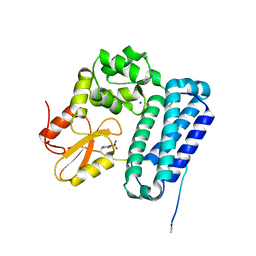 | | Crystal Structure of c-Cbl TKBD in complex with SPRY2 peptide (36-60, pY55) Refined to 1.8 A Resolution (P21 form) | | Descriptor: | E3 ubiquitin-protein ligase CBL, Protein sprouty homolog 2, SODIUM ION | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of c-Cbl TKBD in complex with SPRY2 peptide (36-60, pY55) Refined to 1.8 A Resolution (P21 form)
To be published
|
|
5HKW
 
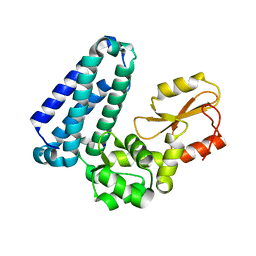 | | Crystal Structure of Apo c-Cbl TKBD Refined to 2.25 A Resolution | | Descriptor: | E3 ubiquitin-protein ligase CBL, SODIUM ION | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Apo c-Cbl TKBD Refined to 2.25 A Resolution
To be published
|
|
8C18
 
 | | Solution structure of carotenoid-binding protein AstaPo1 in complex with astaxanthin | | Descriptor: | ASTAXANTHIN, Astaxanthin binding fasciclin family protein | | Authors: | Kornilov, F.D, Savitskaya, A.G, Slonimskiy, Y.B, Goncharuk, S.A, Sluchanko, N.N, Mineev, K.S. | | Deposit date: | 2022-12-20 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the ligand promiscuity of the neofunctionalized, carotenoid-binding fasciclin domain protein AstaP.
Commun Biol, 6, 2023
|
|
5H94
 
 | | Crystal structure of Swine MHC CLASSI for 1.48 angstroms | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, Nonapeptide from Influenza A virus HA protein | | Authors: | Fan, S, Zhang, N, Wang, S, Wu, Y, Xia, C. | | Deposit date: | 2015-12-25 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural and Biochemical Analyses of Swine Major Histocompatibility Complex Class I Complexes and Prediction of the Epitope Map of Important Influenza A Virus Strains
J.Virol., 90, 2016
|
|
7BV3
 
 | | Crystal structure of a ugt transferase from Siraitia grosvenorii in complex with UDP | | Descriptor: | Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Li, J, Shan, N, Yang, J.G, Liu, W.D, Sun, Y.X. | | Deposit date: | 2020-04-09 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Near-perfect control of the regioselective glucosylation enabled by rational design of glycosyltransferases
Green Synth Catal, 2021
|
|
1PVZ
 
 | | Solution Structure of BmP07, A Novel Potassium Channel Blocker from Scorpion Buthus martensi Karsch, 15 structures | | Descriptor: | K+ toxin-like peptide | | Authors: | Wu, H, Zhang, N, Wang, Y, Zhang, Q, Ou, L, Li, M, Hu, G. | | Deposit date: | 2003-06-29 | | Release date: | 2004-05-18 | | Last modified: | 2018-06-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BmKK2, a new potassium channel blocker from the venom of chinese scorpion Buthus martensi Karsch
PROTEINS, 55, 2004
|
|
7TIV
 
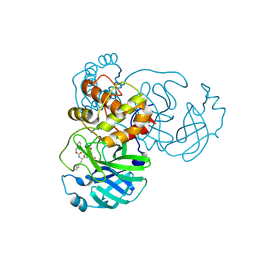 | | Crystal structure of SARS-CoV-2 3CL in complex with inhibitor EB48 | | Descriptor: | (1S,2S)-2-[(N-{[(3-chlorophenyl)methoxy]carbonyl}-3-cyclohexyl-L-alanyl)amino]-1-hydroxy-3-[(3R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, MAGNESIUM ION | | Authors: | Forouhar, F, Liu, H, Iketani, S, Zack, A, Khanizeman, N, Bednarova, E, Fowler, B, Hong, S.J, Mohri, H, Nair, M.S, Huang, Y, Tay, N.E.S, Lee, S, Karan, C, Resnick, S.J, Quinn, C, Li, W, Shion, H, Jurtschenko, C, Lauber, M.A, McDonald, T, Stokes, M.E, Hurst, B, Rovis, T, Chavez, A, Ho, D.D, Stockwell, B.R. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19.
Nat Commun, 13, 2022
|
|
7TIY
 
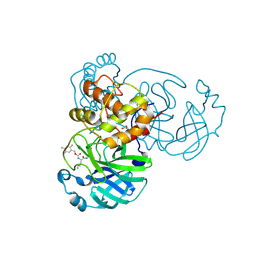 | | Crystal structure of SARS-CoV-2 3CL in complex with inhibitor NK01-48 | | Descriptor: | (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-[(N-{[(2,4,5-trifluorophenyl)methoxy]carbonyl}-L-leucyl)amino]propane-1-sulfonic acid, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | Authors: | Forouhar, F, Liu, H, Iketani, S, Zack, A, Khanizeman, N, Bednarova, E, Fowler, B, Hong, S.J, Mohri, H, Nair, M.S, Huang, Y, Tay, N.E.S, Lee, S, Karan, C, Resnick, S.J, Quinn, C, Li, W, Shion, H, Jurtschenko, C, Lauber, M.A, McDonald, T, Stokes, M.E, Hurst, B, Rovis, T, Chavez, A, Ho, D.D, Stockwell, B.R. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19.
Nat Commun, 13, 2022
|
|
7TIU
 
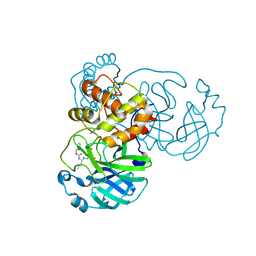 | | Crystal structure of SARS-CoV-2 3CL in complex with inhibitor EB46 | | Descriptor: | (1S,2S)-2-[(N-{[(3-chlorophenyl)methoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, MAGNESIUM ION, ... | | Authors: | Forouhar, F, Liu, H, Iketani, S, Zack, A, Khanizeman, N, Bednarova, E, Fowler, B, Hong, S.J, Mohri, H, Nair, M.S, Huang, Y, Tay, N.E.S, Lee, S, Karan, C, Resnick, S.J, Quinn, C, Li, W, Shion, H, Jurtschenko, C, Lauber, M.A, McDonald, T, Stokes, M.E, Hurst, B, Rovis, T, Chavez, A, Ho, D.D, Stockwell, B.R. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19.
Nat Commun, 13, 2022
|
|
7TIZ
 
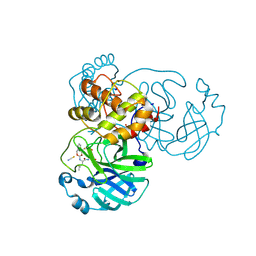 | | Crystal structure of SARS-CoV-2 3CL in complex with inhibitor NK01-63 | | Descriptor: | (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[3-(trifluoromethyl)phenyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | Authors: | Forouhar, F, Liu, H, Iketani, S, Zack, A, Khanizeman, N, Bednarova, E, Fowler, B, Hong, S.J, Mohri, H, Nair, M.S, Huang, Y, Tay, N.E.S, Lee, S, Karan, C, Resnick, S.J, Quinn, C, Li, W, Shion, H, Jurtschenko, C, Lauber, M.A, McDonald, T, Stokes, M.E, Hurst, B, Rovis, T, Chavez, A, Ho, D.D, Stockwell, B.R. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19.
Nat Commun, 13, 2022
|
|
7TIW
 
 | | Crystal structure of SARS-CoV-2 3CL in complex with inhibitor EB54 | | Descriptor: | (1S,2S)-2-[(N-{[(2-chlorophenyl)methoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, PHOSPHATE ION | | Authors: | Forouhar, F, Liu, H, Iketani, S, Zack, A, Khanizeman, N, Bednarova, E, Fowler, B, Hong, S.J, Mohri, H, Nair, M.S, Huang, Y, Tay, N.E.S, Lee, S, Karan, C, Resnick, S.J, Quinn, C, Li, W, Shion, H, Jurtschenko, C, Lauber, M.A, McDonald, T, Stokes, M.E, Hurst, B, Rovis, T, Chavez, A, Ho, D.D, Stockwell, B.R. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19.
Nat Commun, 13, 2022
|
|
