1AK1
 
 | | FERROCHELATASE FROM BACILLUS SUBTILIS | | Descriptor: | FERROCHELATASE | | Authors: | Al-Karadaghi, S, Hansson, M, Nikonov, S, Jonsson, B, Hederstedt, L. | | Deposit date: | 1997-05-28 | | Release date: | 1997-12-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of ferrochelatase: the terminal enzyme in heme biosynthesis.
Structure, 5, 1997
|
|
3UT4
 
 | | Structural view of a non Pfam singleton and crystal packing analysis | | Descriptor: | Putative uncharacterized protein | | Authors: | Cheng, C, Shaw, N, Zhang, X, Zhang, M, Ding, W, Wang, B.C, Liu, Z.J. | | Deposit date: | 2011-11-25 | | Release date: | 2012-03-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural view of a non pfam singleton and crystal packing analysis.
Plos One, 7, 2012
|
|
3C5K
 
 | | Crystal structure of human HDAC6 zinc finger domain | | Descriptor: | Histone deacetylase 6, ZINC ION | | Authors: | Dong, A, Ravichandran, M, Schuetz, A, Loppnau, P, Li, Y, MacKenzie, F, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Dhe-Paganon, S, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-31 | | Release date: | 2008-02-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of Human HDAC6 zinc finger domain.
To be Published
|
|
3CY6
 
 | | Crystal Structure of E18Q DJ-1 | | Descriptor: | Protein DJ-1 | | Authors: | Witt, A.C, Lakshminarasimhan, M, Remington, B.C, Hasim, S, Pozharski, E, Wilson, M.A. | | Deposit date: | 2008-04-25 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Cysteine pKa depression by a protonated glutamic acid in human DJ-1.
Biochemistry, 47, 2008
|
|
3DM1
 
 | | Crystal structure of the complex of human chromobox homolog 3 (CBX3) with peptide | | Descriptor: | Chromobox protein homolog 3, Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 | | Authors: | Amaya, M.F, Ravichandran, M, Loppnau, P, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-06-30 | | Release date: | 2008-08-19 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of the chromodomain of Cbx3 bound to methylated peptides from histone h1 and G9a.
Plos One, 7, 2012
|
|
3UT7
 
 | | Structural view of a non Pfam singleton and crystal packing analysis | | Descriptor: | Putative uncharacterized protein, SULFATE ION | | Authors: | Cheng, C, Shaw, N, Zhang, X, Zhang, M, Ding, W, Wang, B.C, Liu, Z.J. | | Deposit date: | 2011-11-25 | | Release date: | 2012-03-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural view of a non pfam singleton and crystal packing analysis.
Plos One, 7, 2012
|
|
2N3D
 
 | | Atomic structure of the cytoskeletal bactofilin BacA revealed by solid-state NMR | | Descriptor: | Bactofilin A | | Authors: | Shi, C, Fricke, P, Lin, L, Chevelkov, V, Wegstroth, M, Giller, K, Becker, S, Thanbichler, M, Lange, A. | | Deposit date: | 2015-05-29 | | Release date: | 2015-12-16 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic-resolution structure of cytoskeletal bactofilin by solid-state NMR.
Sci Adv, 1, 2015
|
|
2X31
 
 | | Modelling of the complex between subunits BchI and BchD of magnesium chelatase based on single-particle cryo-EM reconstruction at 7.5 ang | | Descriptor: | MAGNESIUM-CHELATASE 38 KDA SUBUNIT, MAGNESIUM-CHELATASE 60 KDA SUBUNIT | | Authors: | Lunqvist, J, Elmlund, H, Peterson Wulff, R, Berglund, L, Elmlund, D, Emanuelsson, C, Hebert, H, Willows, R.D, Hansson, M, Lindahl, M, Al-Karadaghi, S. | | Deposit date: | 2010-01-19 | | Release date: | 2010-11-10 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | ATP-Induced Conformational Dynamics in the Aaa+ Motor Unit of Magnesium Chelatase.
Structure, 18, 2010
|
|
3UT8
 
 | | Structural view of a non Pfam singleton and crystal packing analysis | | Descriptor: | IODIDE ION, Putative uncharacterized protein | | Authors: | Cheng, C, Shaw, N, Zhang, X, Zhang, M, Ding, W, Wang, B.C, Liu, Z.J. | | Deposit date: | 2011-11-25 | | Release date: | 2012-03-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.168 Å) | | Cite: | Structural view of a non pfam singleton and crystal packing analysis.
Plos One, 7, 2012
|
|
3CZ9
 
 | | Crystal Structure of E18L DJ-1 | | Descriptor: | O-ACETALDEHYDYL-HEXAETHYLENE GLYCOL, Protein DJ-1 | | Authors: | Witt, A.C, Lakshminarasimhan, M, Remington, B.C, Hasim, S, Pozharski, E, Wilson, M.A. | | Deposit date: | 2008-04-28 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Cysteine pKa depression by a protonated glutamic acid in human DJ-1.
Biochemistry, 47, 2008
|
|
3CYF
 
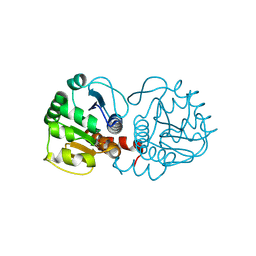 | | Crystal Structure of E18N DJ-1 | | Descriptor: | Protein DJ-1 | | Authors: | Witt, A.C, Lakshminarasimhan, M, Remington, B.C, Hasim, S, Pozharski, E, Wilson, M.A. | | Deposit date: | 2008-04-25 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cysteine pKa depression by a protonated glutamic acid in human DJ-1.
Biochemistry, 47, 2008
|
|
2BSL
 
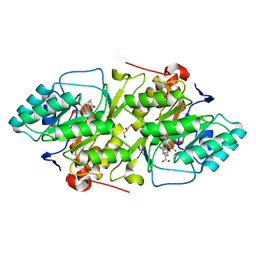 | | Crystal structure of L. lactis dihydroorotate dehydrogense A in complex with 3,4-dihydroxybenzoate | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, ACETATE ION, DIHYDROOROTATE DEHYDROGENASE A, ... | | Authors: | Wolfe, A.E, Hansen, M, Gattis, S.G, Hu, Y.-C, Johansson, E, Arent, S, Larsen, S, Palfey, B.A. | | Deposit date: | 2005-05-23 | | Release date: | 2006-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Interaction of Benzoate Pyrimidine Analogues with Class 1A Dihydroorotate Dehydrogenase from Lactococcus Lactis.
Biochemistry, 46, 2007
|
|
2BX7
 
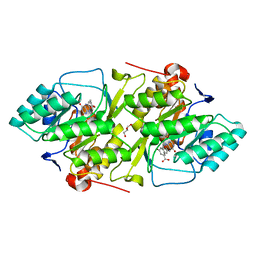 | | Crystal structure of L. lactis dihydroorotate dehydrogense A in complex with 3,5-dihydroxybenzoate | | Descriptor: | 3,5-DIHYDROXYBENZOATE, ACETATE ION, DIHYDROOROTATE DEHYDROGENASE, ... | | Authors: | Wolfe, A.E, Hansen, M, Gattis, S.G, Hu, Y.-C, Johansson, E, Arent, S, Larsen, S, Palfey, B.A. | | Deposit date: | 2005-07-25 | | Release date: | 2006-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Interaction of Benzoate Pyrimidine Analogues with Class 1A Dihydroorotate Dehydrogenase from Lactococcus Lactis.
Biochemistry, 46, 2007
|
|
1EQJ
 
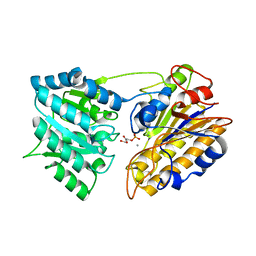 | | CRYSTAL STRUCTURE OF PHOSPHOGLYCERATE MUTASE FROM BACILLUS STEAROTHERMOPHILUS COMPLEXED WITH 2-PHOSPHOGLYCERATE | | Descriptor: | 2-PHOSPHOGLYCERIC ACID, MANGANESE (II) ION, PHOSPHOGLYCERATE MUTASE | | Authors: | Jedrzejas, M.J, Chander, M, Setlow, P, Krishnasamy, G. | | Deposit date: | 2000-04-05 | | Release date: | 2001-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of catalysis of the cofactor-independent phosphoglycerate mutase from Bacillus stearothermophilus. Crystal structure of the complex with 2-phosphoglycerate.
J.Biol.Chem., 275, 2000
|
|
1EJJ
 
 | | CRYSTAL STRUCTURAL ANALYSIS OF PHOSPHOGLYCERATE MUTASE COCRYSTALLIZED WITH 3-PHOSPHOGLYCERATE | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, MANGANESE (II) ION, PHOSPHOGLYCERATE MUTASE | | Authors: | Jedrzejas, M.J, Chander, M, Setlow, P, Krishnasamy, G. | | Deposit date: | 2000-03-02 | | Release date: | 2001-03-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of action of a novel phosphoglycerate mutase from Bacillus stearothermophilus.
EMBO J., 19, 2000
|
|
2OQ0
 
 | | Crystal Structure of the First HIN-200 Domain of Interferon-Inducible Protein 16 | | Descriptor: | CHLORIDE ION, Gamma-interferon-inducible protein Ifi-16 | | Authors: | Lam, R, Liao, J.C.C, Ravichandran, M, Ma, J, Tempel, W, Chirgadze, N.Y, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-01-30 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the First HIN-200 Domain of Interferon-Inducible Protein 16
To be Published
|
|
3U6E
 
 | | MutM set 1 TpGo | | Descriptor: | DNA (5'-D(*A*GP*GP*TP*AP*GP*AP*TP*CP*CP*AP*GP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*CP*TP*(8OG)P*GP*AP*(08Q)P*CP*TP*AP*CP*C)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Sung, R.J, Zhang, M, Qi, Y, Verdine, G.L. | | Deposit date: | 2011-10-12 | | Release date: | 2012-04-25 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Sequence-dependent structural variation in DNA undergoing intrahelical inspection by the DNA glycosylase MutM.
J.Biol.Chem., 287, 2012
|
|
3U6P
 
 | | MutM set 1 GpG | | Descriptor: | DNA (5'-D(*A*GP*GP*TP*AP*GP*AP*TP*CP*CP*CP*GP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*CP*GP*GP*GP*AP*(08Q)P*CP*TP*AP*CP*C)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Sung, R.J, Zhang, M, Qi, Y, Verdine, G.L. | | Deposit date: | 2011-10-12 | | Release date: | 2012-04-25 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Sequence-dependent structural variation in DNA undergoing intrahelical inspection by the DNA glycosylase MutM.
J.Biol.Chem., 287, 2012
|
|
4AV8
 
 | | Kluyveromyces lactis Hsv2 complete loop 6CD | | Descriptor: | SVP1-LIKE PROTEIN 2 | | Authors: | Krick, R, Busse, R.A, Scacioc, A, Stephan, M, Janshoff, A, Thumm, M, Kuhnel, K. | | Deposit date: | 2012-05-24 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structural and Functional Characterization of the Two Phosphoinositide Binding Sites of Proppins, a Beta-Propeller Protein Family.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3U6O
 
 | | MutM set 1 ApG | | Descriptor: | DNA (5'-D(*A*GP*GP*TP*AP*GP*AP*TP*CP*CP*TP*GP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*CP*AP*GP*GP*AP*(08Q)P*CP*TP*AP*CP*C)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Sung, R.J, Zhang, M, Qi, Y, Verdine, G.L. | | Deposit date: | 2011-10-12 | | Release date: | 2012-04-25 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sequence-dependent structural variation in DNA undergoing intrahelical inspection by the DNA glycosylase MutM.
J.Biol.Chem., 287, 2012
|
|
4ER8
 
 | | Structure of the REP associates tyrosine transposase bound to a REP hairpin | | Descriptor: | DNA (32-MER), NICKEL (II) ION, TnpArep for protein | | Authors: | Messing, S.A.J, Ton-Hoang, B, Hickman, A.B, Ghirlando, R, Chandler, M, Dyda, F. | | Deposit date: | 2012-04-19 | | Release date: | 2012-08-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The processing of repetitive extragenic palindromes: the structure of a repetitive extragenic palindrome bound to its associated nuclease.
Nucleic Acids Res., 40, 2012
|
|
3U6L
 
 | | MutM set 2 CpGo | | Descriptor: | DNA (5'-D(*AP*GP*GP*TP*AP*GP*AP*CP*CP*GP*GP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*CP*CP*(8OG)P*GP*TP*(CX2)P*TP*AP*CP*C)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Sung, R.J, Zhang, M, Qi, Y, Verdine, G.L. | | Deposit date: | 2011-10-12 | | Release date: | 2012-04-25 | | Last modified: | 2017-01-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Sequence-dependent structural variation in DNA undergoing intrahelical inspection by the DNA glycosylase MutM.
J.Biol.Chem., 287, 2012
|
|
4AV9
 
 | | Kluyveromyces lactis Hsv2 | | Descriptor: | SULFATE ION, SVP1-LIKE PROTEIN 2 | | Authors: | Krick, R, Busse, R.A, Scacioc, A, Stephan, M, Janshoff, A, Thumm, M, Kuhnel, K. | | Deposit date: | 2012-05-24 | | Release date: | 2012-06-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Structural and Functional Characterization of the Two Phosphoinositide Binding Sites of Proppins, a Beta-Propeller Protein Family.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5OQM
 
 | | STRUCTURE OF YEAST TRANSCRIPTION PRE-INITIATION COMPLEX WITH TFIIH AND CORE MEDIATOR | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Schilbach, S, Hantsche, M, Tegunov, D, Dienemann, C, Wigge, C, Urlaub, H, Cramer, P. | | Deposit date: | 2017-08-13 | | Release date: | 2018-05-09 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structures of transcription pre-initiation complex with TFIIH and Mediator.
Nature, 551, 2017
|
|
3U6M
 
 | | Structural effects of sequence context on lesion recognition by MutM | | Descriptor: | DNA (5'-D(*AP*GP*GP*TP*AP*GP*AP*CP*CP*AP*GP*GP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*CP*CP*TP*(8OG)P*GP*TP*(CX2)P*TP*AP*CP*C)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Sung, R.J, Zhang, M, Qi, Y, Verdine, G.L. | | Deposit date: | 2011-10-12 | | Release date: | 2012-04-25 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Sequence-dependent structural variation in DNA undergoing intrahelical inspection by the DNA glycosylase MutM.
J.Biol.Chem., 287, 2012
|
|
