6OYC
 
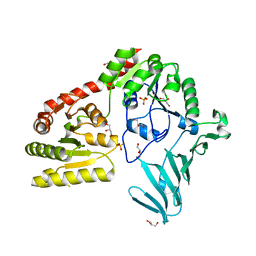 | |
1C1J
 
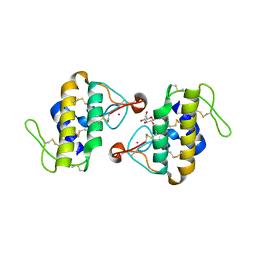 | | STRUCTURE OF CADMIUM-SUBSTITUTED PHOSPHOLIPASE A2 FROM AGKISTRONDON HALYS PALLAS AT 2.8 ANGSTROMS RESOLUTION | | Descriptor: | BASIC PHOSPHOLIPASE A2, CADMIUM ION, octyl beta-D-glucopyranoside | | Authors: | Zhang, H.-l, Zhang, Y.-q, Song, S.-y, Zhou, Y, Lin, Z.-j. | | Deposit date: | 1999-07-22 | | Release date: | 2002-07-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Cadmium-substituted Phospholipase A2 from Agkistrodon halys
Pallas at 2.8 Angstroms Resolution
Protein Pept.Lett., 6, 1999
|
|
8Z4L
 
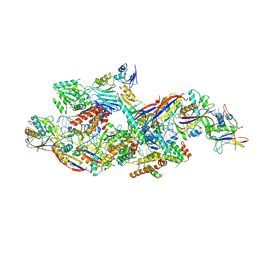 | |
8Z9E
 
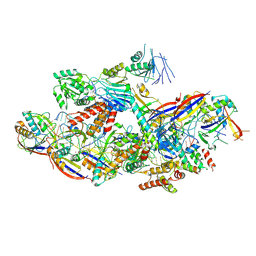 | |
8Z9C
 
 | |
8Z99
 
 | |
8Z4J
 
 | |
8YHE
 
 | |
8YHD
 
 | |
4X2A
 
 | | Crystal structure of mouse glyoxalase I complexed with baicalein | | Descriptor: | 5,6,7-trihydroxy-2-phenyl-4H-chromen-4-one, Lactoylglutathione lyase, ZINC ION | | Authors: | Zhang, H, Zhai, J, Zhang, L, Li, C, Zhao, Y, Hu, X. | | Deposit date: | 2014-11-26 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | In Vitro Inhibition of Glyoxalase І by Flavonoids: New Insights from Crystallographic Analysis.
Curr Top Med Chem, 16, 2016
|
|
8FK2
 
 | | The N-terminal VicR from Streptococcus mutans | | Descriptor: | Putative response regulator CovR VicR-like protein | | Authors: | Zhang, H, Wu, H. | | Deposit date: | 2022-12-20 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Small Molecule Attenuates Bacterial Virulence by Targeting Conserved Response Regulator.
Mbio, 14, 2023
|
|
8GS4
 
 | | Cryo-EM structure of human Neuroligin 2 | | Descriptor: | Neuroligin-2 | | Authors: | Zhang, H, Zhang, Z, Hou, M. | | Deposit date: | 2022-09-04 | | Release date: | 2023-03-01 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Expression and structural analysis of human neuroligin 2 and neuroligin 3 implicated in autism spectrum disorders.
Front Endocrinol, 13, 2022
|
|
5YQZ
 
 | | Structure of the glucagon receptor in complex with a glucagon analogue | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glucagon analogue, ... | | Authors: | Zhang, H, Qiao, A, Yang, L, VAN EPS, N, Frederiksen, K, Yang, D, Dai, A, Cai, X, Zhang, H, Yi, C, Can, C, He, L, Yang, H, Lau, J, Ernst, O, Hanson, M, Stevens, R, Wang, M, Seedtz-Runge, S, Jiang, H, Zhao, Q, Wu, B. | | Deposit date: | 2017-11-08 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the glucagon receptor in complex with a glucagon analogue.
Nature, 553, 2018
|
|
8GS3
 
 | | Cryo-EM structure of human Neuroligin 3 | | Descriptor: | Neuroligin-3 | | Authors: | Zhang, H, Zhang, Z, Hou, M. | | Deposit date: | 2022-09-04 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Expression and structural analysis of human neuroligin 2 and neuroligin 3 implicated in autism spectrum disorders.
Front Endocrinol, 13, 2022
|
|
5HEC
 
 | | CgT structure in dimer | | Descriptor: | Putative glycosyltransferase (GalT1) | | Authors: | Zhang, H, Wu, H. | | Deposit date: | 2016-01-05 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | New Helical Binding Domain Mediates a Glycosyltransferase Activity of a Bifunctional Protein.
J.Biol.Chem., 291, 2016
|
|
7YLT
 
 | |
7YLS
 
 | | Structure of a bacteria protein complex | | Descriptor: | ACETATE ION, Aromatic-ring-hydroxylating dioxygenase beta subunit, FE (III) ION, ... | | Authors: | Zhang, H, Ma, Y.J, Yu, G.M, Li, X.Z. | | Deposit date: | 2022-07-26 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a bacteria protein complex
To Be Published
|
|
7YRJ
 
 | |
7YRI
 
 | |
8GNB
 
 | |
5J39
 
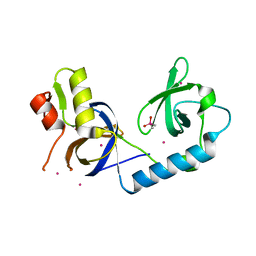 | | Crystal Structure of the extended TUDOR domain from TDRD2 | | Descriptor: | CACODYLATE ION, Tudor and KH domain-containing protein, UNKNOWN ATOM OR ION | | Authors: | Zhang, H, Tempel, W, Dong, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-03-30 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for arginine methylation-independent recognition of PIWIL1 by TDRD2.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7YLR
 
 | |
5E09
 
 | |
5F1K
 
 | | human CD38 in complex with nanobody MU1053 | | Descriptor: | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, nanobody MU1053 | | Authors: | Zhang, H, Hao, Q. | | Deposit date: | 2015-11-30 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Immuno-targeting the multifunctional CD38 using nanobody
Sci Rep, 6, 2016
|
|
5J5K
 
 | | CRYSTAL STRUCTURE OF AFMP4P IN COMPLEX WITH PALMITIC ACID | | Descriptor: | PALMITIC ACID, Uncharacterized protein, alpha-D-mannopyranose | | Authors: | Zhang, H, Hao, Q. | | Deposit date: | 2016-04-03 | | Release date: | 2017-04-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Novel Class Of Virulence Factors In Penicillium Marneffei And Aspergillus Fumigatus Enhances Intracellular Survival In Monocytes By Arachidonic Acid Binding
To Be Published
|
|
