4J4Y
 
 | | Triple mutant GraVN | | Descriptor: | NP protein | | Authors: | Jiao, L, Ouyang, S, Liang, M, Niu, F, Shaw, N, Wu, W, Ding, W, Jin, C, Zhu, Y, Zhang, F, Wang, T, Li, C, Zuo, X, Luan, C.H, Li, D, Liu, Z.J. | | Deposit date: | 2013-02-07 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of severe Fever with thrombocytopenia syndrome virus nucleocapsid protein in complex with suramin reveals therapeutic potential
J.Virol., 87, 2013
|
|
4J4U
 
 | | Pentamer SFTSVN | | Descriptor: | Nucleocapsid protein | | Authors: | Jiao, L, Ouyang, S, Liang, M, Niu, F, Shaw, N, Wu, W, Ding, W, Jin, C, Zhu, Y, Zhang, F, Wang, T, Li, C, Zuo, X, Luan, C.H, Li, D, Liu, Z.J. | | Deposit date: | 2013-02-07 | | Release date: | 2013-05-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Structure of severe Fever with thrombocytopenia syndrome virus nucleocapsid protein in complex with suramin reveals therapeutic potential
J.Virol., 87, 2013
|
|
4YM5
 
 | | Crystal structure of the human nucleosome containing 6-4PP (inside) | | Descriptor: | 144 mer-DNA, 144-mer DNA, Histone H2A type 1-B/E, ... | | Authors: | Osakabe, A, Tachiwana, H, Kagawa, W, Horikoshi, N, Matsumoto, S, Hasegawa, M, Matsumoto, N, Toga, T, Yamamoto, J, Hanaoka, F, Thoma, N.H, Sugasawa, K, Iwai, S, Kurumizaka, H. | | Deposit date: | 2015-03-06 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.005 Å) | | Cite: | Structural basis of pyrimidine-pyrimidone (6-4) photoproduct recognition by UV-DDB in the nucleosome
Sci Rep, 5, 2015
|
|
9C0J
 
 | |
9C0I
 
 | |
9CEU
 
 | | Spizellomyces punctatus Fanzor (SpuFz) State 1 | | Descriptor: | DNA (5'-D(P*CP*CP*TP*AP*TP*AP*GP*AP*TP*AP*TP*GP*CP*CP*CP*GP*GP*GP*TP*AP*CP*CP*G)-3'), DNA (5'-D(P*CP*GP*GP*TP*AP*CP*CP*CP*GP*GP*GP*CP*AP*TP*A)-3'), Maltose/maltodextrin-binding periplasmic protein,Spizellomyces punctatus Fanzor 1, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CEV
 
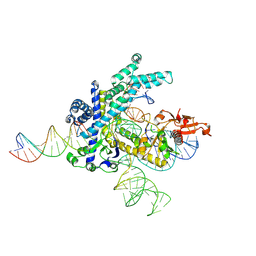 | | Spizellomyces punctatus Fanzor (SpuFz) State 2 | | Descriptor: | DNA (35-MER), DNA (5'-D(P*CP*GP*GP*TP*AP*CP*CP*CP*GP*GP*GP*CP*AP*TP*A)-3'), MAGNESIUM ION, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CEY
 
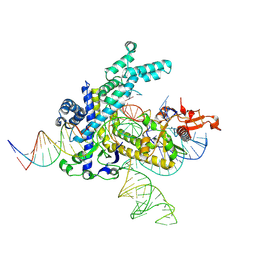 | | Spizellomyces punctatus Fanzor (SpuFz) State 5 | | Descriptor: | DNA (26-MER), DNA (36-MER), MAGNESIUM ION, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CF3
 
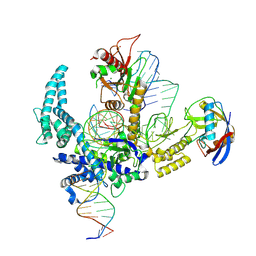 | | Parasitella parasitica Fanzor (PpFz) State 4 | | Descriptor: | DNA non-target strand, DNA target strand, MAGNESIUM ION, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CER
 
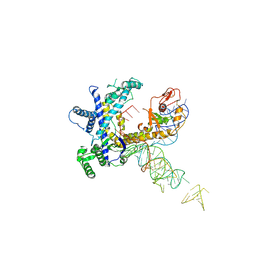 | | Guillardia theta Fanzor (GtFz) State 1 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Guillardia theta Fanzor1, RNA (142-MER), ZINC ION | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CF2
 
 | | Parasitella parasitica Fanzor (PpFz) State 3 | | Descriptor: | DNA non-target strand, DNA substrate model, DNA target strand, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CEZ
 
 | | Spizellomyces punctatus Fanzor (SpuFz) State 6 | | Descriptor: | DNA (27-MER), DNA (5'-D(P*TP*AP*CP*CP*CP*GP*GP*GP*CP*AP*TP*A)-3'), MAGNESIUM ION, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CEW
 
 | | Spizellomyces punctatus Fanzor (SpuFz) State 3 | | Descriptor: | DNA (29-MER), DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*CP*GP*GP*TP*AP*CP*CP*CP*GP*GP*GP*CP*AP*TP*A)-3'), ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CF0
 
 | | Parasitella parasitica Fanzor (PpFz) State 1 | | Descriptor: | DNA non-target strand, DNA target strand, Maltose/maltodextrin-binding periplasmic protein,Parasitella parasitica Fanzor 1, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CEX
 
 | | Spizellomyces punctatus Fanzor (SpuFz) State 4 | | Descriptor: | DNA (29-MER), DNA (5'-D(*(MG)*(MG)P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*CP*GP*GP*TP*AP*CP*CP*CP*GP*GP*GP*CP*AP*TP*A)-3'), ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CET
 
 | | Guillardia theta Fanzor (GtFz) State 3 | | Descriptor: | DNA (28-MER), DNA (5'-D(P*AP*TP*GP*AP*CP*TP*TP*CP*TP*CP*TP*TP*AP*AP*AP*GP*GP*CP*CP*CP*CP*GP*GP*G)-3'), Maltose/maltodextrin-binding periplasmic protein,Guillardia theta Fanzor1, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CF1
 
 | | Parasitella parasitica Fanzor (PpFz) State 2 | | Descriptor: | DNA non-target strand, DNA target strand, Maltose/maltodextrin-binding periplasmic protein,Parasitella parasitica Fanzor 1, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CES
 
 | | Guillardia theta Fanzor (GtFz) State 2 | | Descriptor: | DNA (5'-D(P*AP*TP*GP*AP*CP*TP*TP*CP*TP*CP*TP*TP*AP*AP*AP*GP*GP*CP*CP*CP*CP*GP*GP*G)-3'), DNA (5'-D(P*CP*CP*CP*GP*GP*GP*GP*CP*CP*TP*TP*TP*AP*AP*G)-3'), Maltose/maltodextrin-binding periplasmic protein,Guillardia theta Fanzor1, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
4J4X
 
 | | Crystal structure of GraVN | | Descriptor: | NP protein | | Authors: | Jiao, L, Ouyang, S, Liang, M, Niu, F, Shaw, N, Wu, W, Ding, W, Jin, C, Zhu, Y, Zhang, F, Wang, T, Li, C, Zuo, X, Luan, C.H, Li, D, Liu, Z.J. | | Deposit date: | 2013-02-07 | | Release date: | 2013-05-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure of severe Fever with thrombocytopenia syndrome virus nucleocapsid protein in complex with suramin reveals therapeutic potential
J.Virol., 87, 2013
|
|
4J4W
 
 | | Crystal structure of BueVN | | Descriptor: | Nucleocapsid | | Authors: | Jiao, L, Ouyang, S, Liang, M, Niu, F, Shaw, N, Wu, W, Ding, W, Jin, C, Zhu, Y, Zhang, F, Wang, T, Li, C, Zuo, X, Luan, C.H, Li, D, Liu, Z.J. | | Deposit date: | 2013-02-07 | | Release date: | 2013-05-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.661 Å) | | Cite: | Structure of severe Fever with thrombocytopenia syndrome virus nucleocapsid protein in complex with suramin reveals therapeutic potential
J.Virol., 87, 2013
|
|
3H4P
 
 | | Proteasome 20S core particle from Methanocaldococcus jannaschii | | Descriptor: | Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Jeffrey, P.D, Zhang, F, Hu, M, Tian, G, Zhang, P, Finley, D, Shi, Y. | | Deposit date: | 2009-04-20 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structural Insights into the Regulatory Particle of the Proteasome from Methanocaldococcus jannaschii.
Mol.Cell, 34, 2009
|
|
3H4M
 
 | | AAA ATPase domain of the proteasome- activating nucleotidase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Proteasome-activating nucleotidase | | Authors: | Jeffrey, P, Zhang, F, Hu, M, Tian, G, Zhang, P, Finley, D, Shi, Y. | | Deposit date: | 2009-04-20 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.106 Å) | | Cite: | Structural Insights into the Regulatory Particle of the Proteasome from Methanocaldococcus jannaschii.
Mol.Cell, 34, 2009
|
|
3H43
 
 | | N-terminal domain of the proteasome-activating nucleotidase of Methanocaldococcus jannaschii | | Descriptor: | Proteasome-activating nucleotidase | | Authors: | Jeffrey, P.D, Zhang, F, Hu, M, Tian, G, Zhang, P, Finley, D, Shi, Y. | | Deposit date: | 2009-04-17 | | Release date: | 2009-06-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insights into the Regulatory Particle of the Proteasome from Methanocaldococcus jannaschii.
Mol.Cell, 34, 2009
|
|
1RY7
 
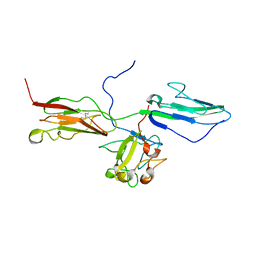 | | Crystal Structure of the 3 Ig form of FGFR3c in complex with FGF1 | | Descriptor: | Fibroblast growth factor receptor 3, Heparin-binding growth factor 1 | | Authors: | Olsen, S.K, Ibrahimi, O.A, Raucci, A, Zhang, F, Eliseenkova, A.V, Yayon, A, Basilico, C, Linhardt, R.J, Schlessinger, J, Mohammadi, M. | | Deposit date: | 2003-12-19 | | Release date: | 2004-02-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Insights into the molecular basis for fibroblast growth factor receptor autoinhibition and ligand-binding promiscuity.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
3CYJ
 
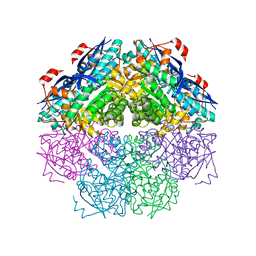 | | Crystal structure of a mandelate racemase/muconate lactonizing enzyme-like protein from Rubrobacter xylanophilus | | Descriptor: | GLYCEROL, Mandelate racemase/muconate lactonizing enzyme-like protein, SODIUM ION | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Zhang, F, Bravo, J, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-25 | | Release date: | 2008-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a mandelate racemase/muconate lactonizing enzyme-like protein from Rubrobacter xylanophilus.
To be Published
|
|
