4BNH
 
 | | Crystal structure of S. aureus FabI in complex with NADP and 5-hexyl- 2-phenoxyphenol | | Descriptor: | 5-hexyl-2-phenoxyphenol, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], GLUTAMIC ACID, ... | | Authors: | Schiebel, J, Chang, A, Bommineni, G.R, Tonge, P.J, Kisker, C. | | Deposit date: | 2013-05-15 | | Release date: | 2013-06-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Rational Optimization of Drug-Target Residence Time: Insights from Inhibitor Binding to the S. Aureus Fabi Enzyme-Product Complex.
Biochemistry, 52, 2013
|
|
4BNM
 
 | | Crystal structure of S. aureus FabI in complex with NADP and 5-hexyl- 2-(2-methylphenoxy)phenol | | Descriptor: | 5-HEXYL-2-(2-METHYLPHENOXY)PHENOL, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], GLUTAMIC ACID, ... | | Authors: | Schiebel, J, Chang, A, Bommineni, G.R, Tonge, P.J, Kisker, C. | | Deposit date: | 2013-05-15 | | Release date: | 2013-06-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Rational Optimization of Drug-Target Residence Time: Insights from Inhibitor Binding to the S. Aureus Fabi Enzyme-Product Complex.
Biochemistry, 52, 2013
|
|
4BNF
 
 | | Crystal structure of S. aureus FabI in complex with NADP and 2- phenoxy-5-propylphenol | | Descriptor: | 2-phenoxy-5-propyl-phenol, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], GLUTAMIC ACID, ... | | Authors: | Schiebel, J, Chang, A, Bommineni, G.R, Tonge, P.J, Kisker, C. | | Deposit date: | 2013-05-15 | | Release date: | 2013-06-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational Optimization of Drug-Target Residence Time: Insights from Inhibitor Binding to the S. Aureus Fabi Enzyme-Product Complex.
Biochemistry, 52, 2013
|
|
7LH8
 
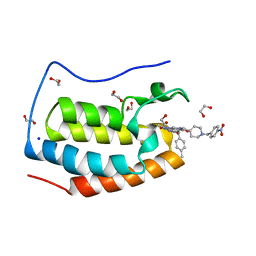 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH INHIBITOR JJ-II-131 | | Descriptor: | (5R)-1-ethyl-5-(4-methylphenyl)-7-({1-[(4-nitrophenyl)methyl]piperidin-4-yl}methyl)-5,7,8,9-tetrahydro-1H-pyrrolo[3',4':5,6]pyrido[2,3-d]pyrimidine-2,4,6(3H)-trione, 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ... | | Authors: | Schonbrunn, E, Chan, A. | | Deposit date: | 2021-01-21 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Dihydropyridine Lactam Analogs Targeting BET Bromodomains.
Chemmedchem, 17, 2022
|
|
4BNJ
 
 | | Crystal structure of S. aureus FabI in complex with NADP and 5-methyl- 2-phenoxyphenol | | Descriptor: | 5-methyl-2-phenoxyphenol, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], GLUTAMIC ACID, ... | | Authors: | Schiebel, J, Chang, A, Bommineni, G.R, Tonge, P.J, Kisker, C. | | Deposit date: | 2013-05-15 | | Release date: | 2013-06-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Rational Optimization of Drug-Target Residence Time: Insights from Inhibitor Binding to the S. Aureus Fabi Enzyme-Product Complex.
Biochemistry, 52, 2013
|
|
8Q68
 
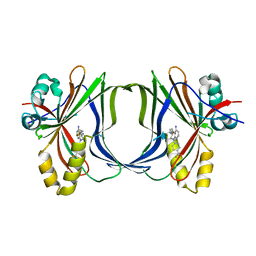 | | Crystal structure of TEAD1-YBD in complex with irreversible compound SWTX-143 | | Descriptor: | Transcriptional enhancer factor TEF-1, ~{N}-[(3~{S})-5-azanyl-1-[4-(trifluoromethyl)phenyl]-3,4-dihydro-2~{H}-quinolin-3-yl]propanamide | | Authors: | Ciesielski, F, Spieser, S.A.H, Marchand, A, Gwaltney, S.L. | | Deposit date: | 2023-08-11 | | Release date: | 2023-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | A Novel Irreversible TEAD Inhibitor, SWTX-143, Blocks Hippo Pathway Transcriptional Output and Causes Tumor Regression in Preclinical Mesothelioma Models.
Mol.Cancer Ther., 23, 2024
|
|
4BNI
 
 | | Crystal structure of S. aureus FabI in complex with NADP and 2-(2- aminophenoxy)-5-hexylphenol | | Descriptor: | 2-(2-azanylphenoxy)-5-hexyl-phenol, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], GLUTAMIC ACID, ... | | Authors: | Schiebel, J, Chang, A, Bommineni, G.R, Tonge, P.J, Kisker, C. | | Deposit date: | 2013-05-15 | | Release date: | 2013-06-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational Optimization of Drug-Target Residence Time: Insights from Inhibitor Binding to the S. Aureus Fabi Enzyme-Product Complex.
Biochemistry, 52, 2013
|
|
8B6X
 
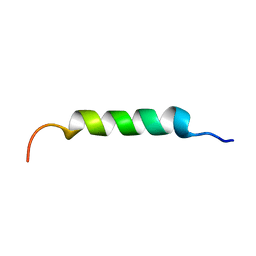 | |
8B6Y
 
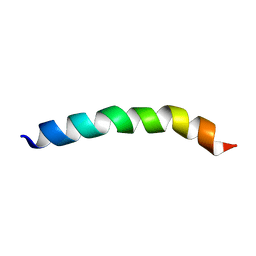 | |
1VRH
 
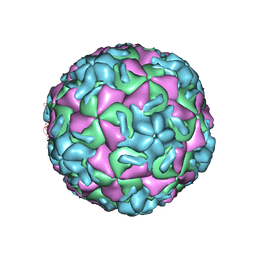 | | HRV14/SDZ 880-061 COMPLEX | | Descriptor: | 2-[4-(2H-1,4-BENZOTHIAZINE-3-YL)-PIPERAZINE-1-LY]-1,3-THIAZOLE-4-CARBOXYLIC ACID ETHYLESTER, RHINOVIRUS 14 | | Authors: | Oren, D.A, Zhang, A, Arnold, E. | | Deposit date: | 1996-02-26 | | Release date: | 1997-02-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Synthesis and activity of piperazine-containing antirhinoviral agents and crystal structure of SDZ 880-061 bound to human rhinovirus 14.
J.Mol.Biol., 259, 1996
|
|
4BNK
 
 | | Crystal structure of S. aureus FabI in complex with NADP and 5-fluoro- 2-phenoxyphenol | | Descriptor: | 5-fluoro-2-phenoxyphenol, CHLORIDE ION, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], ... | | Authors: | Schiebel, J, Chang, A, Bommineni, G.R, Tonge, P.J, Kisker, C. | | Deposit date: | 2013-05-15 | | Release date: | 2013-06-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rational Optimization of Drug-Target Residence Time: Insights from Inhibitor Binding to the S. Aureus Fabi Enzyme-Product Complex.
Biochemistry, 52, 2013
|
|
4BNG
 
 | | Crystal structure of S. aureus FabI in complex with NADP and 5-pentyl- 2-phenoxyphenol | | Descriptor: | 5-PENTYL-2-PHENOXYPHENOL, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], GLUTAMIC ACID, ... | | Authors: | Schiebel, J, Chang, A, Bommineni, G.R, Tonge, P.J, Kisker, C. | | Deposit date: | 2013-05-15 | | Release date: | 2013-06-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational Optimization of Drug-Target Residence Time: Insights from Inhibitor Binding to the S. Aureus Fabi Enzyme-Product Complex.
Biochemistry, 52, 2013
|
|
4BNN
 
 | | Crystal structure of S. aureus FabI in complex with NADP and 2-(2- cyanophenoxy)-5-hexylphenol | | Descriptor: | 2-(2-CYANOPHENOXY)-5-HEXYLPHENOL, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], GLUTAMIC ACID, ... | | Authors: | Schiebel, J, Chang, A, Bommineni, G.R, Tonge, P.J, Kisker, C. | | Deposit date: | 2013-05-15 | | Release date: | 2013-06-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Rational Optimization of Drug-Target Residence Time: Insights from Inhibitor Binding to the S. Aureus Fabi Enzyme-Product Complex.
Biochemistry, 52, 2013
|
|
4CV0
 
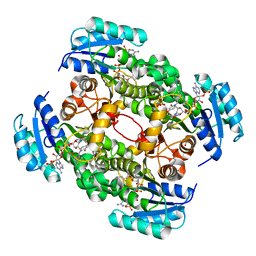 | | Crystal structure of S. aureus FabI in complex with NADPH and CG400549 (small unit cell) | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-amino-2-methylbenzyl)-4-[2-(thiophen-2-yl)ethoxy]pyridin-2(1H)-one, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], ... | | Authors: | Schiebel, J, Chang, A, Shah, S, Tonge, P.J, Kisker, C. | | Deposit date: | 2014-03-22 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational Design of Broad Spectrum Antibacterial Activity Based on a Clinically Relevant Enoyl-Acyl Carrier Protein (Acp) Reductase Inhibitor.
J.Biol.Chem., 289, 2014
|
|
4D46
 
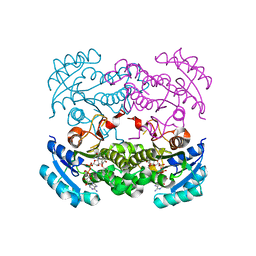 | | Crystal structure of E. coli FabI in complex with NAD and 5-bromo-2-(4-chloro-2-hydroxyphenoxy)benzonitrile | | Descriptor: | 5-bromo-2-(4-chloro-2-hydroxyphenoxy)benzonitrile, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Tareilus, M, Schiebel, J, Chang, A, Tonge, P.J, Sotriffer, C.A, Kisker, C. | | Deposit date: | 2014-10-26 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An Ordered Water Channel in Staphylococcus Aureus Fabi: Unraveling the Mechanism of Substrate Recognition and Reduction.
Biochemistry, 54, 2015
|
|
4CV2
 
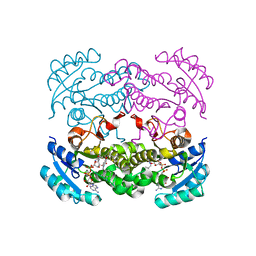 | | Crystal structure of E. coli FabI in complex with NADH and CG400549 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 1-(3-amino-2-methylbenzyl)-4-[2-(thiophen-2-yl)ethoxy]pyridin-2(1H)-one, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH] | | Authors: | Tareilus, M, Schiebel, J, Chang, A, Shah, S, Tonge, P.J, Kisker, C. | | Deposit date: | 2014-03-22 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rational Design of Broad Spectrum Antibacterial Activity Based on a Clinically Relevant Enoyl-Acyl Carrier Protein (Acp) Reductase Inhibitor.
J.Biol.Chem., 289, 2014
|
|
6LF7
 
 | | Crystal structure of the complex of goat lactoperoxidase with hypothiocyanite and hydrogen peroxide at 1.79 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Viswanathan, V, Tyagi, T.K, Singh, R.P, Singh, A.K, Singh, A, Bhushan, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2019-11-30 | | Release date: | 2020-01-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.794 Å) | | Cite: | Crystal structure of the complex of goat lactoperoxidase with hypothiocyanite and hydrogen peroxide at 1.79 A resolution.
To Be Published
|
|
5AB8
 
 | | High resolution X-ray structure of the N-terminal truncated form (residues 1-11) of Mycobacterium tuberculosis HbN | | Descriptor: | ACETATE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Pesce, A, Bustamante, J.P, Bidon-Chanal, A, Boechi, L, Estrin, D.A, Luque, F.J, Sebilo, A, Guertin, M, Bolognesi, M, Ascenzi, P, Nardini, M. | | Deposit date: | 2015-08-04 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | The N-Terminal Pre-A Region of Mycobacterium Tuberculosis 2/2Hbn Promotes No-Dioxygenase Activity.
FEBS J., 283, 2016
|
|
6S13
 
 | | Erythromycin Resistant Staphylococcus aureus 70S ribosome (delta R88 A89 uL22). | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Halfon, Y, Matozv, D, Eyal, Z, Bashan, A, Zimmerman, E, Kjeldgaard, J, Ingmer, H, Yonath, A. | | Deposit date: | 2019-06-18 | | Release date: | 2019-08-21 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Exit tunnel modulation as resistance mechanism of S. aureus erythromycin resistant mutant.
Sci Rep, 9, 2019
|
|
6S12
 
 | | Erythromycin Resistant Staphylococcus aureus 50S ribosome (delta R88 A89 uL22). | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Halfon, Y, Matozv, D, Eyal, Z, Bashan, A, Zimmerman, E, Kjeldgaard, J, Ingmer, H, Yonath, A. | | Deposit date: | 2019-06-18 | | Release date: | 2019-08-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Exit tunnel modulation as resistance mechanism of S. aureus erythromycin resistant mutant.
Sci Rep, 9, 2019
|
|
5TCU
 
 | | Methicillin sensitive Staphylococcus aureus 70S ribosome | | Descriptor: | 16S RRNA, 23S RRNA, 30S ribosomal protein S10, ... | | Authors: | Eyal, Z, Ahmed, T, Belousoff, N, Mishra, S, Matzov, D, Bashan, A, Zimmerman, E, Lithgow, T, Bhushan, S, Yonath, A. | | Deposit date: | 2016-09-15 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Basis for Linezolid Binding Site Rearrangement in the Staphylococcus aureus Ribosome.
MBio, 8, 2017
|
|
4GOW
 
 | | Crystal Structure of Ca2+/CaM:Kv7.4 (KCNQ4) B helix complex | | Descriptor: | CALCIUM ION, Calmodulin, Potassium voltage-gated channel subfamily KQT member 4 | | Authors: | Xu, Q, Chang, A, Tolia, A, Minor Jr, D.L. | | Deposit date: | 2012-08-20 | | Release date: | 2012-12-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of a Ca(2+)/CaM:Kv7.4 (KCNQ4) B-helix complex provides insight into M current modulation.
J.Mol.Biol., 425, 2013
|
|
4D43
 
 | | Crystal structure of S. aureus FabI in complex with NADP and 2-(2- chloro-4-nitrophenoxy)-5-ethyl-4-fluorophenol | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-(2-chloro-4-nitrophenoxy)-5-ethyl-4-fluorophenol, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], ... | | Authors: | Schiebel, J, Chang, A, Tonge, P.J, Sotriffer, C.A, Kisker, C. | | Deposit date: | 2014-10-26 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | An Ordered Water Channel in Staphylococcus Aureus Fabi: Unraveling the Mechanism of Substrate Recognition and Reduction.
Biochemistry, 54, 2015
|
|
4D44
 
 | | Crystal structure of S. aureus FabI in complex with NADP and 5-ethyl- 4-fluoro-2-((2-fluoropyridin-3-yl)oxy)phenol | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 5-ethyl-4-fluoro-2-[(2-fluoropyridin-3-yl)oxy]phenol, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], ... | | Authors: | Schiebel, J, Chang, A, Tonge, P.J, Sotriffer, C.A, Kisker, C. | | Deposit date: | 2014-10-26 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Ordered Water Channel in Staphylococcus Aureus Fabi: Unraveling the Mechanism of Substrate Recognition and Reduction.
Biochemistry, 54, 2015
|
|
3LY5
 
 | | DDX18 dead-domain | | Descriptor: | ATP-dependent RNA helicase DDX18, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Schutz, P, Karlberg, T, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Kraulis, P, Kotenyova, T, Kotzsch, A, Markova, N, Moche, M, Nielsen, T.K, Nordlund, P, Nyman, T, Persson, C, Roos, A.K, Siponen, M.I, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-02-26 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Comparative Structural Analysis of Human DEAD-Box RNA Helicases.
Plos One, 5, 2010
|
|
